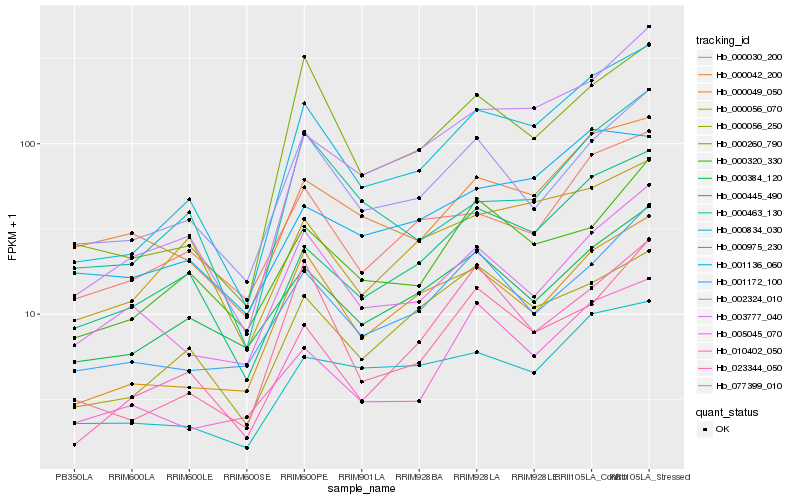
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000975_230 |
0.0 |
- |
- |
hypothetical protein ZEAMMB73_864543 [Zea mays] |
| 2 |
Hb_000445_490 |
0.098639649 |
- |
- |
- |
| 3 |
Hb_003777_040 |
0.1030805518 |
- |
- |
PREDICTED: sm-like protein LSM3A [Jatropha curcas] |
| 4 |
Hb_000049_050 |
0.1068250557 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 5 |
Hb_000030_200 |
0.1213770186 |
- |
- |
PREDICTED: early nodulin-93-like [Populus euphratica] |
| 6 |
Hb_000260_790 |
0.1223214536 |
- |
- |
NADH dehydrogenase 1 alpha subcomplex subunit 12 isoform 1 [Theobroma cacao] |
| 7 |
Hb_000834_030 |
0.1289113458 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000042_200 |
0.1296646227 |
- |
- |
PREDICTED: uncharacterized protein LOC100252136 isoform X1 [Vitis vinifera] |
| 9 |
Hb_001172_100 |
0.1298373083 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000463_130 |
0.1307636604 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_002324_010 |
0.1307945865 |
- |
- |
PREDICTED: proteasome subunit alpha type-7 [Jatropha curcas] |
| 12 |
Hb_005045_070 |
0.1326256982 |
- |
- |
PREDICTED: deSI-like protein At4g17486 [Jatropha curcas] |
| 13 |
Hb_000384_120 |
0.1331260732 |
transcription factor |
TF Family: Whirly |
PREDICTED: single-stranded DNA-bindig protein WHY2, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000056_250 |
0.1333835646 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein 3 [Jatropha curcas] |
| 15 |
Hb_010402_050 |
0.1338336333 |
- |
- |
- |
| 16 |
Hb_077399_010 |
0.1348862177 |
- |
- |
hypothetical protein POPTR_0003s09910g [Populus trichocarpa] |
| 17 |
Hb_001136_060 |
0.1351634399 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 18 |
Hb_023344_050 |
0.1360376588 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 19 |
Hb_000056_070 |
0.1363412345 |
- |
- |
- |
| 20 |
Hb_000320_330 |
0.1372990806 |
- |
- |
uncharacterized protein LOC100527884 [Glycine max] |