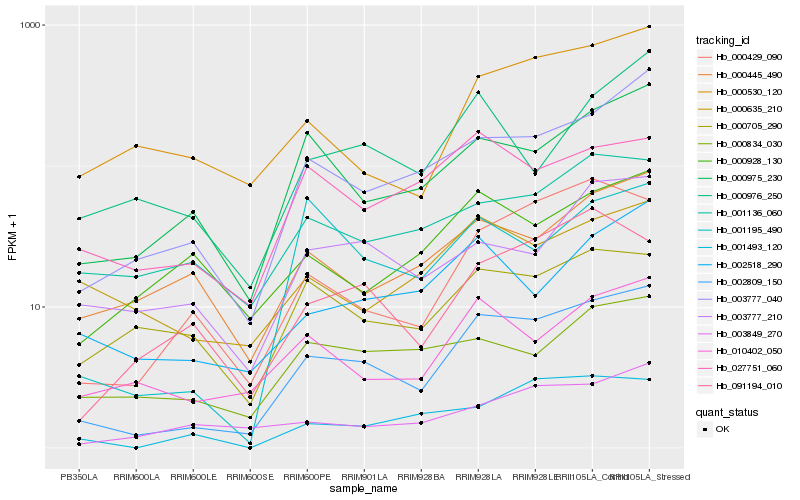
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002809_150 |
0.0 |
transcription factor |
TF Family: M-type |
hypothetical protein CISIN_1g0283632mg, partial [Citrus sinensis] |
| 2 |
Hb_003777_040 |
0.1393050391 |
- |
- |
PREDICTED: sm-like protein LSM3A [Jatropha curcas] |
| 3 |
Hb_000429_090 |
0.1547208989 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000975_230 |
0.1616462986 |
- |
- |
hypothetical protein ZEAMMB73_864543 [Zea mays] |
| 5 |
Hb_010402_050 |
0.1717954382 |
- |
- |
- |
| 6 |
Hb_002518_290 |
0.179627914 |
- |
- |
Acyl-coenzyme A thioesterase 13 [Gossypium arboreum] |
| 7 |
Hb_001493_120 |
0.1805356711 |
- |
- |
- |
| 8 |
Hb_000530_120 |
0.1812678997 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 9 |
Hb_091194_010 |
0.1817803023 |
- |
- |
hypothetical protein POPTR_0006s05430g [Populus trichocarpa] |
| 10 |
Hb_001195_490 |
0.1847951494 |
- |
- |
hypothetical protein glysoja_018593 [Glycine soja] |
| 11 |
Hb_000445_490 |
0.1876001423 |
- |
- |
- |
| 12 |
Hb_001136_060 |
0.1881477451 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 13 |
Hb_003777_210 |
0.1887109349 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 14 |
Hb_003849_270 |
0.190981059 |
- |
- |
- |
| 15 |
Hb_027751_060 |
0.1928871189 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B [Jatropha curcas] |
| 16 |
Hb_000976_250 |
0.196848779 |
- |
- |
- |
| 17 |
Hb_000834_030 |
0.1996596552 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000705_290 |
0.1997391806 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |
| 19 |
Hb_000635_210 |
0.2011651371 |
- |
- |
unknown [Populus trichocarpa] |
| 20 |
Hb_000928_130 |
0.2014630821 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |