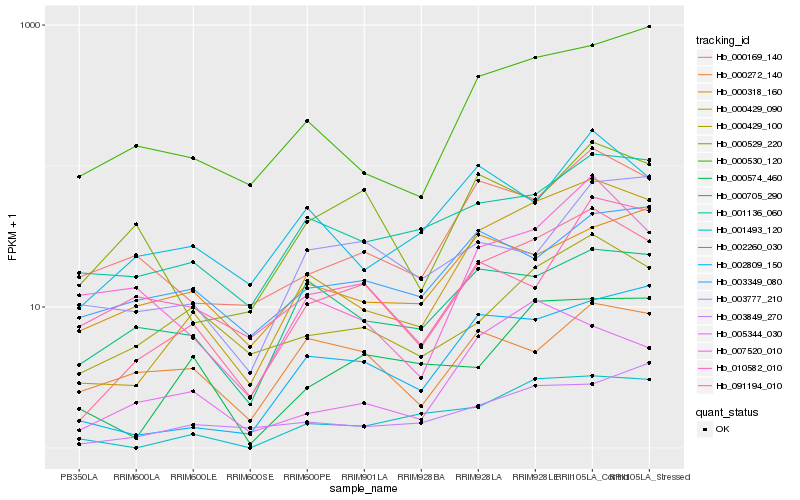
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_091194_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s05430g [Populus trichocarpa] |
| 2 |
Hb_000429_090 |
0.1177377106 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000429_100 |
0.1677703514 |
- |
- |
hypothetical protein POPTR_0006s05430g [Populus trichocarpa] |
| 4 |
Hb_000169_140 |
0.1764643925 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 5 |
Hb_001136_060 |
0.1774898516 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 6 |
Hb_002809_150 |
0.1817803023 |
transcription factor |
TF Family: M-type |
hypothetical protein CISIN_1g0283632mg, partial [Citrus sinensis] |
| 7 |
Hb_000705_290 |
0.1859921079 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |
| 8 |
Hb_000529_220 |
0.1913929146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000574_460 |
0.1925152241 |
- |
- |
hypothetical protein JCGZ_06119 [Jatropha curcas] |
| 10 |
Hb_002260_030 |
0.1961651456 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 11 |
Hb_000530_120 |
0.1983684183 |
- |
- |
copper transport protein ATOX1 [Hevea brasiliensis] |
| 12 |
Hb_001493_120 |
0.1984501899 |
- |
- |
- |
| 13 |
Hb_003849_270 |
0.1989432303 |
- |
- |
- |
| 14 |
Hb_003777_210 |
0.2035545267 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 15 |
Hb_003349_080 |
0.2055631376 |
- |
- |
PREDICTED: uncharacterized protein LOC105628287 [Jatropha curcas] |
| 16 |
Hb_000272_140 |
0.2059368811 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 17 |
Hb_007520_010 |
0.2059798473 |
- |
- |
PREDICTED: uncharacterized protein LOC105640383 [Jatropha curcas] |
| 18 |
Hb_005344_030 |
0.2086645652 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Jatropha curcas] |
| 19 |
Hb_010582_010 |
0.2110562717 |
- |
- |
hypothetical protein POPTR_0011s15670g [Populus trichocarpa] |
| 20 |
Hb_000318_160 |
0.2132436005 |
- |
- |
PREDICTED: uncharacterized protein LOC105634239 [Jatropha curcas] |