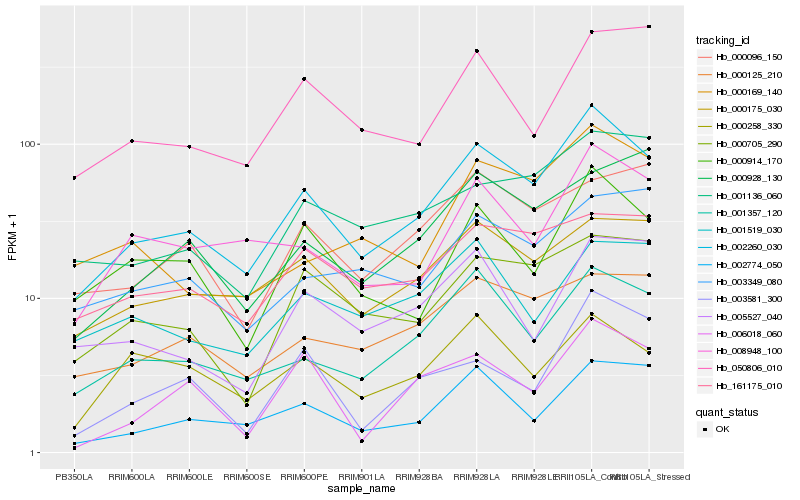
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002260_030 |
0.0 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 2 |
Hb_001357_120 |
0.1182937237 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 3 |
Hb_002774_050 |
0.1364422636 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 4 |
Hb_000914_170 |
0.1385982542 |
- |
- |
hypothetical protein JCGZ_25909 [Jatropha curcas] |
| 5 |
Hb_008948_100 |
0.1395616922 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 6 |
Hb_000175_030 |
0.142412332 |
- |
- |
Signal recognition particle, SRP54 subunit protein [Theobroma cacao] |
| 7 |
Hb_000705_290 |
0.1474818409 |
- |
- |
C-4 methyl sterol oxidase, putative [Ricinus communis] |
| 8 |
Hb_000169_140 |
0.1478809454 |
- |
- |
PREDICTED: uncharacterized protein LOC105643397 [Jatropha curcas] |
| 9 |
Hb_000125_210 |
0.1480950018 |
- |
- |
PREDICTED: electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial [Jatropha curcas] |
| 10 |
Hb_001136_060 |
0.1488625868 |
- |
- |
PREDICTED: uncharacterized protein LOC105641994 [Jatropha curcas] |
| 11 |
Hb_161175_010 |
0.1522885623 |
- |
- |
PREDICTED: vesicle-associated protein 1-1 [Jatropha curcas] |
| 12 |
Hb_050806_010 |
0.158868728 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-17 kDa [Jatropha curcas] |
| 13 |
Hb_005527_040 |
0.158909315 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 14 |
Hb_000096_150 |
0.1607663627 |
- |
- |
NADH-ubiquinone oxidoreductase 18 kDa subunit, mitochondrial precursor, putative [Ricinus communis] |
| 15 |
Hb_003349_080 |
0.1614851778 |
- |
- |
PREDICTED: uncharacterized protein LOC105628287 [Jatropha curcas] |
| 16 |
Hb_001519_030 |
0.163632259 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003581_300 |
0.1638329578 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000928_130 |
0.1672192809 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 19 |
Hb_006018_060 |
0.1672912791 |
- |
- |
Zinc finger SWIM domain-containing protein 7 isoform 1 [Theobroma cacao] |
| 20 |
Hb_000258_330 |
0.1679440544 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |