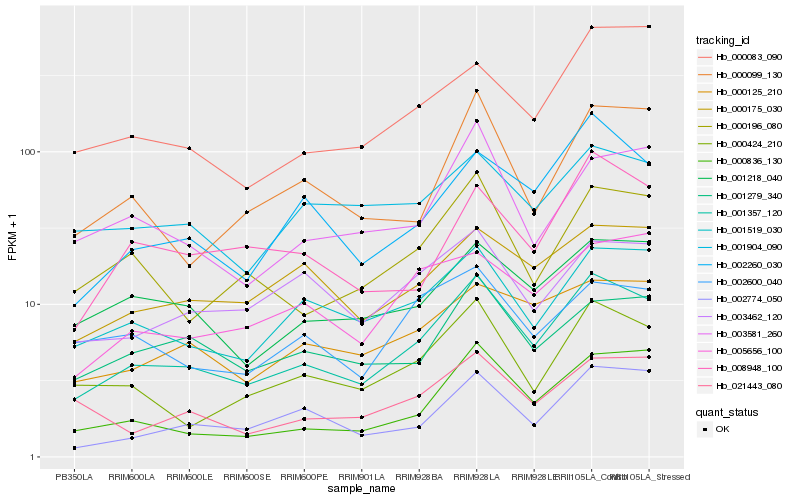
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001357_120 |
0.0 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 2 |
Hb_001519_030 |
0.1131324351 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000836_130 |
0.1178616737 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002260_030 |
0.1182937237 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 5 |
Hb_000424_210 |
0.1184119111 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000196_080 |
0.1223926247 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001218_040 |
0.122741943 |
- |
- |
Mitochondrial substrate carrier family protein [Theobroma cacao] |
| 8 |
Hb_000125_210 |
0.1255987237 |
- |
- |
PREDICTED: electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial [Jatropha curcas] |
| 9 |
Hb_002774_050 |
0.1256089688 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 10 |
Hb_000175_030 |
0.1285379221 |
- |
- |
Signal recognition particle, SRP54 subunit protein [Theobroma cacao] |
| 11 |
Hb_001279_340 |
0.1316038227 |
- |
- |
glycerol-3-phosphate dehydrogenase [Jatropha curcas] |
| 12 |
Hb_005656_100 |
0.1349881865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003581_260 |
0.13610532 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |
| 14 |
Hb_000099_130 |
0.1363172605 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 15 |
Hb_003462_120 |
0.1367382206 |
- |
- |
Acidic endochitinase [Theobroma cacao] |
| 16 |
Hb_008948_100 |
0.1369351152 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 17 |
Hb_000083_090 |
0.1385351254 |
- |
- |
immunophilin, putative [Ricinus communis] |
| 18 |
Hb_002600_040 |
0.1394065684 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 19 |
Hb_001904_090 |
0.1395108882 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 20 |
Hb_021443_080 |
0.1410243706 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |