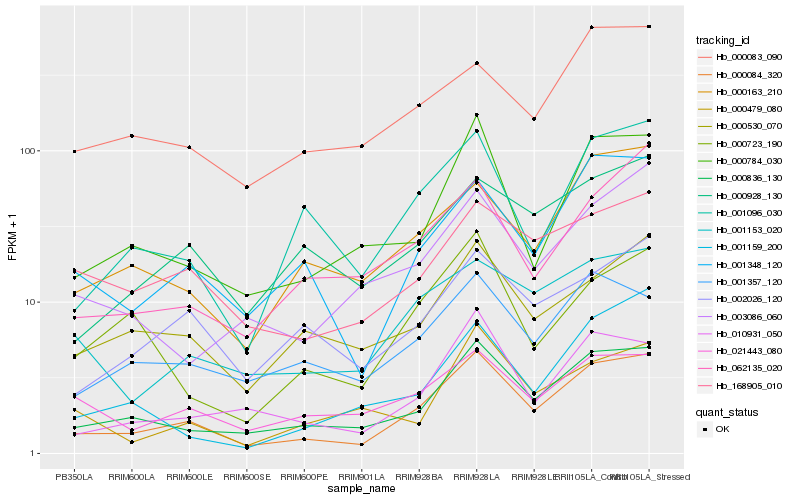
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000084_320 |
0.0 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 2 |
Hb_000836_130 |
0.1084919626 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001348_120 |
0.136198726 |
- |
- |
Phloem protein 2-B10, putative [Theobroma cacao] |
| 4 |
Hb_010931_050 |
0.1423433465 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 5 |
Hb_000784_030 |
0.1444967614 |
- |
- |
neutral/alkaline invertase 2 [Hevea brasiliensis] |
| 6 |
Hb_001153_020 |
0.1522661647 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18-like [Cucumis melo] |
| 7 |
Hb_001096_030 |
0.1525067317 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 8 |
Hb_000163_210 |
0.1559533057 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 9 |
Hb_000530_070 |
0.1614769762 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 10 |
Hb_002026_120 |
0.162769707 |
- |
- |
hypothetical protein CISIN_1g025276mg [Citrus sinensis] |
| 11 |
Hb_001357_120 |
0.1655876483 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 12 |
Hb_168905_010 |
0.1676779231 |
- |
- |
PREDICTED: AP-1 complex subunit sigma-1 isoform X2 [Cicer arietinum] |
| 13 |
Hb_062135_020 |
0.1677836254 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Nicotiana sylvestris] |
| 14 |
Hb_000928_130 |
0.168520928 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 15 |
Hb_003086_060 |
0.1696000962 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 16 |
Hb_000723_190 |
0.1701285937 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001159_200 |
0.1707209952 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_000083_090 |
0.1724294656 |
- |
- |
immunophilin, putative [Ricinus communis] |
| 19 |
Hb_021443_080 |
0.1735288636 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 20 |
Hb_000479_080 |
0.174762292 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |