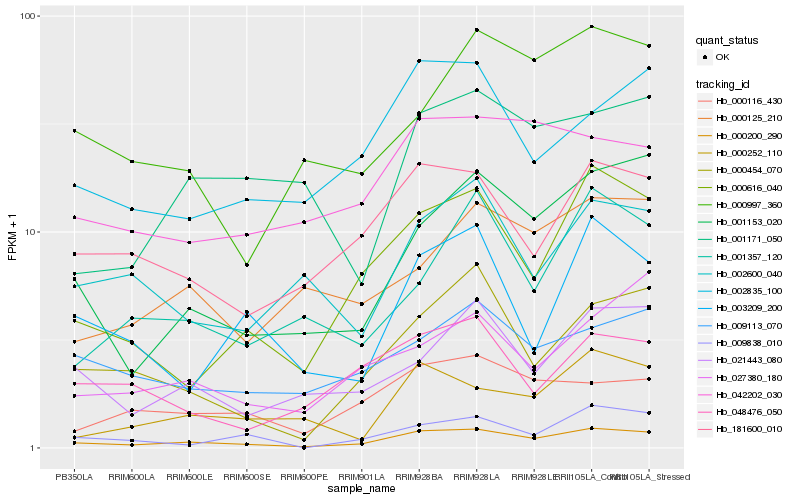
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000200_290 |
0.0 |
- |
- |
PREDICTED: elongator complex protein 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001153_020 |
0.1361829252 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18-like [Cucumis melo] |
| 3 |
Hb_000616_040 |
0.139396357 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 4 |
Hb_000454_070 |
0.1428174516 |
- |
- |
PREDICTED: 30S ribosomal protein S31, mitochondrial [Cucumis sativus] |
| 5 |
Hb_181600_010 |
0.1431415626 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 6 |
Hb_002835_100 |
0.155406935 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 7 |
Hb_021443_080 |
0.1555830089 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 8 |
Hb_000116_430 |
0.1565313874 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 1, mitochondrial-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_009113_070 |
0.1631329417 |
- |
- |
hypothetical protein POPTR_0010s07510g [Populus trichocarpa] |
| 10 |
Hb_009838_010 |
0.1679976347 |
- |
- |
- |
| 11 |
Hb_042202_030 |
0.168754354 |
- |
- |
PREDICTED: uncharacterized protein LOC105647255 [Jatropha curcas] |
| 12 |
Hb_003209_200 |
0.1697619517 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1621230 [Ricinus communis] |
| 13 |
Hb_002600_040 |
0.1711961206 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 14 |
Hb_000252_110 |
0.1722499743 |
- |
- |
- |
| 15 |
Hb_027380_180 |
0.1722800325 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Jatropha curcas] |
| 16 |
Hb_000997_360 |
0.1753655638 |
- |
- |
hypothetical protein AALP_AAs70636U000100 [Arabis alpina] |
| 17 |
Hb_048476_050 |
0.1757017236 |
- |
- |
semialdehyde dehydrogenase family protein [Populus trichocarpa] |
| 18 |
Hb_000125_210 |
0.1776064894 |
- |
- |
PREDICTED: electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001357_120 |
0.1780107046 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 20 |
Hb_001171_050 |
0.1782104796 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0284459 [Jatropha curcas] |