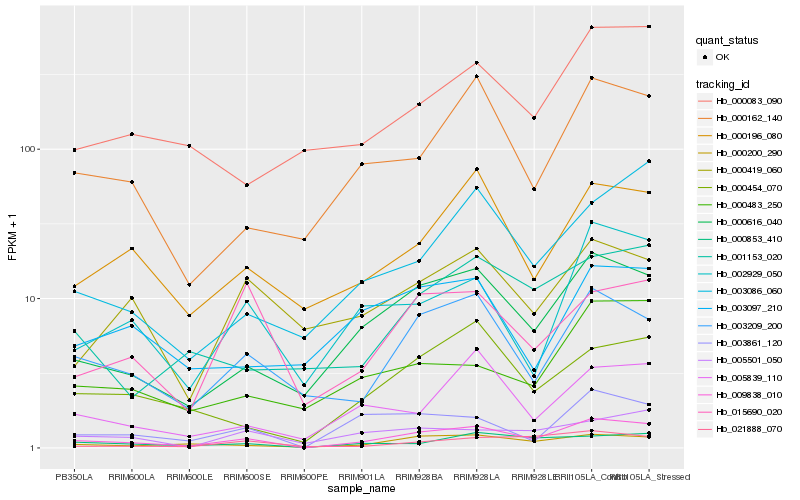
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009838_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000616_040 |
0.1340421804 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 3 |
Hb_003209_200 |
0.1604512033 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1621230 [Ricinus communis] |
| 4 |
Hb_000200_290 |
0.1679976347 |
- |
- |
PREDICTED: elongator complex protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_003861_120 |
0.1771844664 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_000162_140 |
0.177727594 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2 [Jatropha curcas] |
| 7 |
Hb_000196_080 |
0.1781726552 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002929_050 |
0.1800945125 |
- |
- |
PREDICTED: polyadenylate-binding protein 6 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000454_070 |
0.183491029 |
- |
- |
PREDICTED: 30S ribosomal protein S31, mitochondrial [Cucumis sativus] |
| 10 |
Hb_000853_410 |
0.1840500178 |
- |
- |
- |
| 11 |
Hb_005839_110 |
0.1912164803 |
- |
- |
PREDICTED: cytochrome b561 and DOMON domain-containing protein At5g47530-like [Populus euphratica] |
| 12 |
Hb_000483_250 |
0.195661168 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 13 |
Hb_001153_020 |
0.196077975 |
- |
- |
PREDICTED: histone deacetylase complex subunit SAP18-like [Cucumis melo] |
| 14 |
Hb_015690_020 |
0.196397817 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 15 |
Hb_005501_050 |
0.1979689415 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 16 |
Hb_003086_060 |
0.2005185061 |
- |
- |
PREDICTED: uncharacterized protein LOC105629937 [Jatropha curcas] |
| 17 |
Hb_000083_090 |
0.2008325407 |
- |
- |
immunophilin, putative [Ricinus communis] |
| 18 |
Hb_021888_070 |
0.2022481526 |
- |
- |
Putative CCR4-associated factor 1 like 11 [Glycine soja] |
| 19 |
Hb_003097_210 |
0.2039563029 |
- |
- |
PREDICTED: uncharacterized protein LOC105631194 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000419_060 |
0.2044023599 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |