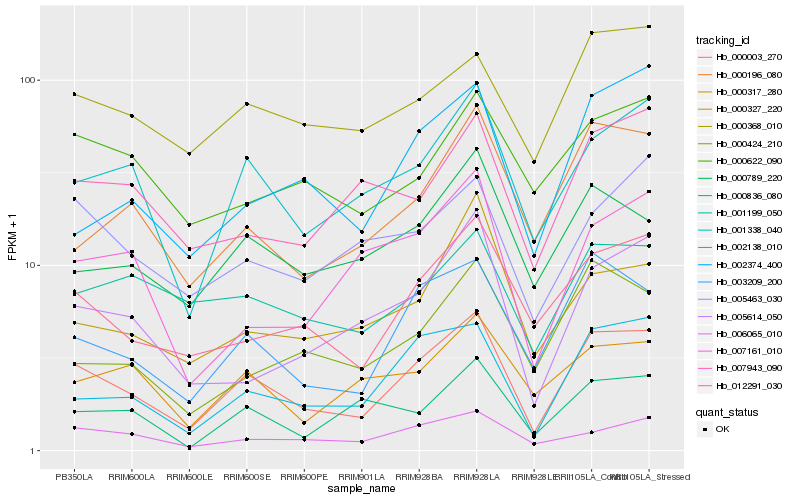
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_270 |
0.0 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 2 |
Hb_003209_200 |
0.1257067897 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1621230 [Ricinus communis] |
| 3 |
Hb_002138_010 |
0.1295795753 |
- |
- |
hypothetical protein POPTR_0001s30620g [Populus trichocarpa] |
| 4 |
Hb_001338_040 |
0.1342907041 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_012291_030 |
0.1364107591 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X3 [Populus euphratica] |
| 6 |
Hb_006065_010 |
0.1433299927 |
- |
- |
PREDICTED: uncharacterized protein LOC105639733 [Jatropha curcas] |
| 7 |
Hb_000196_080 |
0.155275901 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_005614_050 |
0.159468305 |
- |
- |
PREDICTED: aspartate aminotransferase, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_000368_010 |
0.1603522173 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 18 [Jatropha curcas] |
| 10 |
Hb_000789_220 |
0.1647570337 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 11 |
Hb_000424_210 |
0.1673562157 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002374_400 |
0.1680987768 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_005463_030 |
0.168586274 |
- |
- |
PREDICTED: uncharacterized protein LOC105632512 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000317_280 |
0.1703134652 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At5g08490 [Jatropha curcas] |
| 15 |
Hb_000836_080 |
0.1705026428 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g46050, mitochondrial [Jatropha curcas] |
| 16 |
Hb_007943_090 |
0.171091799 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 17 |
Hb_001199_050 |
0.1712761774 |
- |
- |
PREDICTED: DNA-3-methyladenine glycosylase-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000327_220 |
0.1722774718 |
- |
- |
PREDICTED: probable protein phosphatase 2C 38 [Jatropha curcas] |
| 19 |
Hb_007161_010 |
0.1724507481 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000622_090 |
0.1735021742 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |