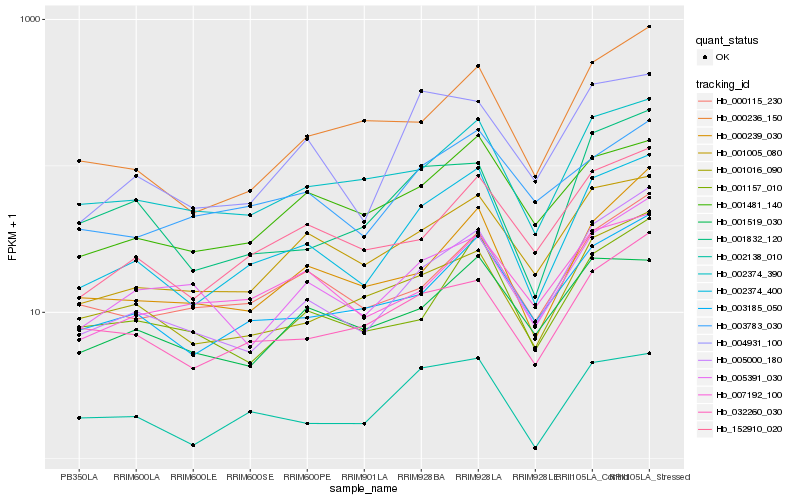
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002374_400 |
0.0 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_003185_050 |
0.1027426108 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 3 |
Hb_005000_180 |
0.1030132307 |
- |
- |
hypothetical protein JCGZ_07408 [Jatropha curcas] |
| 4 |
Hb_002138_010 |
0.1067040029 |
- |
- |
hypothetical protein POPTR_0001s30620g [Populus trichocarpa] |
| 5 |
Hb_002374_390 |
0.1096640427 |
- |
- |
vacuolar ATP synthase proteolipid subunit 1, 2, 3, putative [Ricinus communis] |
| 6 |
Hb_001005_080 |
0.1097354898 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_001016_090 |
0.1161855533 |
- |
- |
PREDICTED: receptor homology region, transmembrane domain- and RING domain-containing protein 1 [Populus euphratica] |
| 8 |
Hb_152910_020 |
0.116816723 |
- |
- |
PREDICTED: lactoylglutathione lyase-like [Jatropha curcas] |
| 9 |
Hb_001481_140 |
0.1195930759 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 10 |
Hb_004931_100 |
0.1213177808 |
- |
- |
Actin depolymerizing factor 6 isoform 1 [Theobroma cacao] |
| 11 |
Hb_032260_030 |
0.1236910613 |
- |
- |
hypothetical protein POPTR_0001s10500g [Populus trichocarpa] |
| 12 |
Hb_001157_010 |
0.1271700669 |
- |
- |
deoxyhypusine synthase, putative [Ricinus communis] |
| 13 |
Hb_000239_030 |
0.1273657299 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 14 |
Hb_000115_230 |
0.1290750725 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 15 |
Hb_003783_030 |
0.1309102587 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |
| 16 |
Hb_000236_150 |
0.1319176799 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_001519_030 |
0.1325414289 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005391_030 |
0.1325441491 |
- |
- |
putative cytidine/deoxycytidylate deaminase family protein [Zea mays] |
| 19 |
Hb_001832_120 |
0.1348450278 |
- |
- |
hypothetical protein PHAVU_003G136800g [Phaseolus vulgaris] |
| 20 |
Hb_007192_100 |
0.1350870286 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |