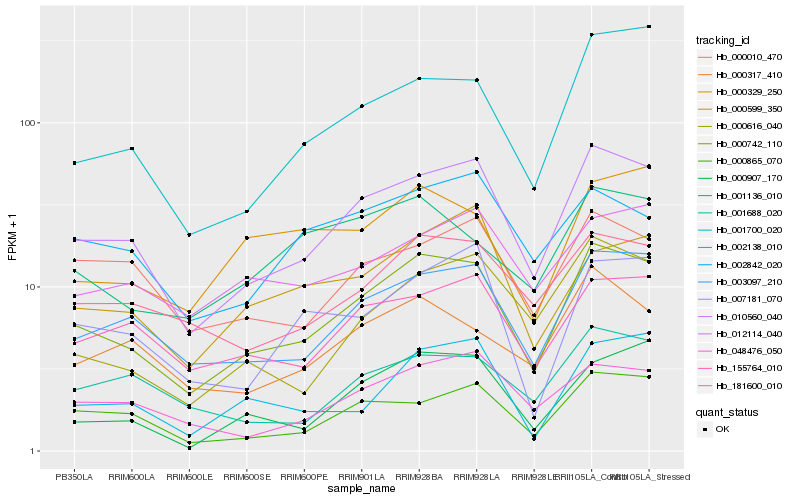
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000742_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_09215 [Jatropha curcas] |
| 2 |
Hb_010560_040 |
0.0639269506 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 11 [Jatropha curcas] |
| 3 |
Hb_003097_210 |
0.0977882392 |
- |
- |
PREDICTED: uncharacterized protein LOC105631194 isoform X1 [Jatropha curcas] |
| 4 |
Hb_181600_010 |
0.1214235538 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 5 |
Hb_155764_010 |
0.127341338 |
- |
- |
- |
| 6 |
Hb_000865_070 |
0.1289289261 |
- |
- |
Protein FAR1-RELATED SEQUENCE 3 [Glycine soja] |
| 7 |
Hb_048476_050 |
0.1337379007 |
- |
- |
semialdehyde dehydrogenase family protein [Populus trichocarpa] |
| 8 |
Hb_001700_020 |
0.133892027 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |
| 9 |
Hb_000907_170 |
0.1346749233 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_007181_070 |
0.1378615357 |
- |
- |
PREDICTED: alpha-galactosidase-like [Jatropha curcas] |
| 11 |
Hb_000317_410 |
0.1394469702 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C5-like [Jatropha curcas] |
| 12 |
Hb_000616_040 |
0.1396492544 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 13 |
Hb_001136_010 |
0.1423256775 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 14 |
Hb_002138_010 |
0.1423301965 |
- |
- |
hypothetical protein POPTR_0001s30620g [Populus trichocarpa] |
| 15 |
Hb_000599_350 |
0.1429035803 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_000329_250 |
0.1435283822 |
- |
- |
PREDICTED: vesicle-associated protein 1-3 isoform X1 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_012114_040 |
0.1452467715 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_000010_470 |
0.1458506434 |
- |
- |
Galactose oxidase/kelch repeat superfamily protein isoform 2 [Theobroma cacao] |
| 19 |
Hb_001688_020 |
0.1466010077 |
- |
- |
PREDICTED: uncharacterized protein LOC103438026 [Malus domestica] |
| 20 |
Hb_002842_020 |
0.1474832433 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |