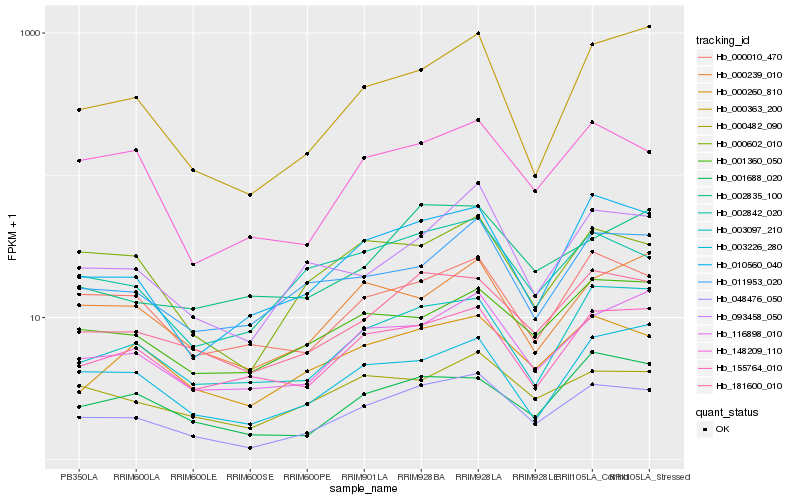
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048476_050 |
0.0 |
- |
- |
semialdehyde dehydrogenase family protein [Populus trichocarpa] |
| 2 |
Hb_181600_010 |
0.0794640398 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 3 |
Hb_116898_010 |
0.1050028026 |
- |
- |
PREDICTED: uncharacterized protein LOC105639569 isoform X2 [Jatropha curcas] |
| 4 |
Hb_155764_010 |
0.1083604484 |
- |
- |
- |
| 5 |
Hb_010560_040 |
0.1085445854 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 11 [Jatropha curcas] |
| 6 |
Hb_000010_470 |
0.1093990452 |
- |
- |
Galactose oxidase/kelch repeat superfamily protein isoform 2 [Theobroma cacao] |
| 7 |
Hb_003097_210 |
0.109735366 |
- |
- |
PREDICTED: uncharacterized protein LOC105631194 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002842_020 |
0.1107945027 |
- |
- |
Prolactin regulatory element-binding protein, putative [Ricinus communis] |
| 9 |
Hb_000482_090 |
0.1115035877 |
- |
- |
PREDICTED: uncharacterized protein LOC105638145 [Jatropha curcas] |
| 10 |
Hb_000260_810 |
0.1116106718 |
- |
- |
- |
| 11 |
Hb_000602_010 |
0.1156577941 |
- |
- |
PREDICTED: rhomboid-like protease 2 isoform X1 [Jatropha curcas] |
| 12 |
Hb_148209_110 |
0.1161144554 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 13 |
Hb_093458_050 |
0.1180755556 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 14 |
Hb_011953_020 |
0.1208437465 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 15 |
Hb_001360_050 |
0.1217756816 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001688_020 |
0.1242071475 |
- |
- |
PREDICTED: uncharacterized protein LOC103438026 [Malus domestica] |
| 17 |
Hb_003226_280 |
0.1269882382 |
- |
- |
PREDICTED: peter Pan-like protein [Populus euphratica] |
| 18 |
Hb_000363_200 |
0.1285329752 |
- |
- |
40S ribosomal protein S8, putative [Ricinus communis] |
| 19 |
Hb_000239_010 |
0.1294813678 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_002835_100 |
0.1307669856 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |