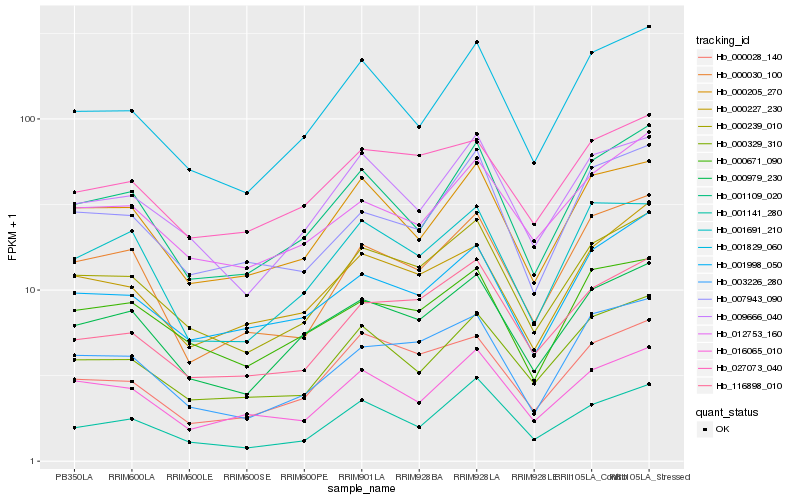
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000239_010 |
0.0 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 2 |
Hb_000979_230 |
0.0613265061 |
- |
- |
PREDICTED: protein BCCIP homolog [Jatropha curcas] |
| 3 |
Hb_009666_040 |
0.0640050846 |
- |
- |
glutathione transferase, putative [Ricinus communis] |
| 4 |
Hb_003226_280 |
0.0671029944 |
- |
- |
PREDICTED: peter Pan-like protein [Populus euphratica] |
| 5 |
Hb_116898_010 |
0.0680242636 |
- |
- |
PREDICTED: uncharacterized protein LOC105639569 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001141_280 |
0.0740578943 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Vitis vinifera] |
| 7 |
Hb_000205_270 |
0.0743391982 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000028_140 |
0.0747981496 |
- |
- |
surfeit locus protein, putative [Ricinus communis] |
| 9 |
Hb_001829_060 |
0.0749405356 |
- |
- |
Eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 10 |
Hb_000329_310 |
0.0759914509 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_027073_040 |
0.0766153218 |
- |
- |
TGF-beta-inducible nuclear protein, putative [Ricinus communis] |
| 12 |
Hb_016065_010 |
0.0769363462 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g08305 [Jatropha curcas] |
| 13 |
Hb_001109_020 |
0.0774222623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000671_090 |
0.0777506411 |
- |
- |
PREDICTED: melanoma-associated antigen 1 [Jatropha curcas] |
| 15 |
Hb_012753_160 |
0.0834864217 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 16 |
Hb_001998_050 |
0.0835233252 |
- |
- |
PREDICTED: alpha N-terminal protein methyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_000227_230 |
0.0836916088 |
- |
- |
PREDICTED: putative methyltransferase C9orf114 [Jatropha curcas] |
| 18 |
Hb_000030_100 |
0.0842678123 |
- |
- |
PREDICTED: short-chain type dehydrogenase/reductase isoform X2 [Jatropha curcas] |
| 19 |
Hb_007943_090 |
0.0859941326 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 20 |
Hb_001691_210 |
0.0880330245 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |