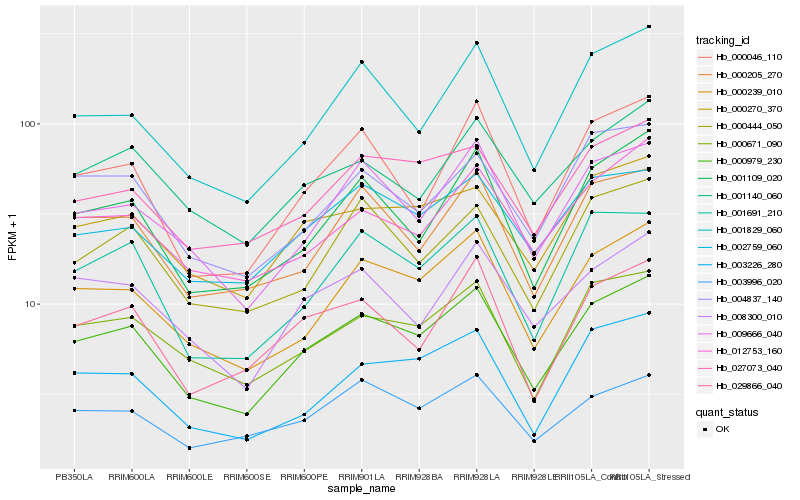
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000979_230 |
0.0 |
- |
- |
PREDICTED: protein BCCIP homolog [Jatropha curcas] |
| 2 |
Hb_000239_010 |
0.0613265061 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 3 |
Hb_000671_090 |
0.0702462203 |
- |
- |
PREDICTED: melanoma-associated antigen 1 [Jatropha curcas] |
| 4 |
Hb_000270_370 |
0.0703418588 |
- |
- |
Uncharacterized protein TCM_011954 [Theobroma cacao] |
| 5 |
Hb_001691_210 |
0.0729228915 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |
| 6 |
Hb_001109_020 |
0.0737142796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009666_040 |
0.0740160556 |
- |
- |
glutathione transferase, putative [Ricinus communis] |
| 8 |
Hb_027073_040 |
0.0771558834 |
- |
- |
TGF-beta-inducible nuclear protein, putative [Ricinus communis] |
| 9 |
Hb_002759_060 |
0.0789283162 |
- |
- |
PREDICTED: protein LSM12 homolog A-like [Jatropha curcas] |
| 10 |
Hb_000205_270 |
0.0791526534 |
- |
- |
PREDICTED: uncharacterized protein LOC105632722 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001829_060 |
0.0793757705 |
- |
- |
Eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 12 |
Hb_003226_280 |
0.0832025264 |
- |
- |
PREDICTED: peter Pan-like protein [Populus euphratica] |
| 13 |
Hb_008300_010 |
0.0837194782 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG13 homolog [Jatropha curcas] |
| 14 |
Hb_001140_060 |
0.0838491608 |
- |
- |
PREDICTED: ADP-ribosylation factor-related protein 1 [Prunus mume] |
| 15 |
Hb_012753_160 |
0.0844739362 |
- |
- |
ara4-interacting protein, putative [Ricinus communis] |
| 16 |
Hb_004837_140 |
0.0845292265 |
- |
- |
hypothetical protein PRUPE_ppa012469mg [Prunus persica] |
| 17 |
Hb_000046_110 |
0.0845439462 |
- |
- |
PREDICTED: 54S ribosomal protein L39, mitochondrial [Jatropha curcas] |
| 18 |
Hb_029866_040 |
0.0850503035 |
- |
- |
PREDICTED: plastid division protein PDV1 [Jatropha curcas] |
| 19 |
Hb_003996_020 |
0.0853851525 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000444_050 |
0.085797026 |
transcription factor |
TF Family: HMG |
PREDICTED: uncharacterized protein LOC105648212 isoform X2 [Jatropha curcas] |