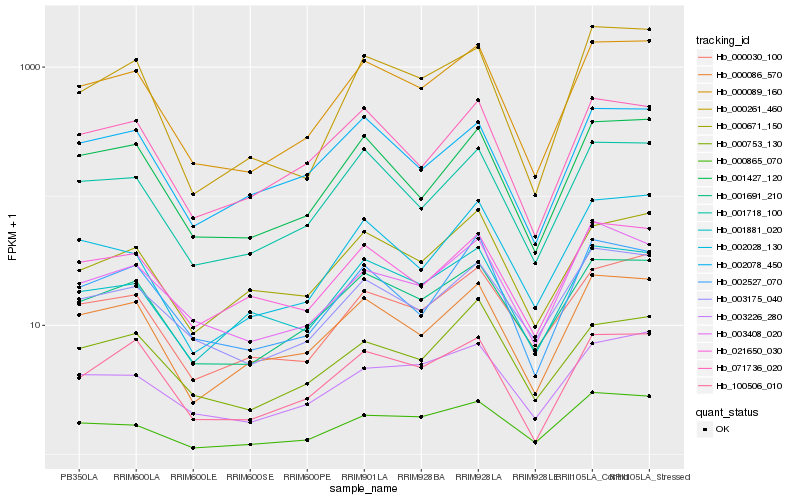
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000089_160 |
0.0 |
- |
- |
60S ribosomal protein L5B [Hevea brasiliensis] |
| 2 |
Hb_001691_210 |
0.0540391557 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |
| 3 |
Hb_001718_100 |
0.0633125692 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 4 |
Hb_000086_570 |
0.0667693573 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 5 |
Hb_001427_120 |
0.0674173387 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 6 |
Hb_000030_100 |
0.0751142001 |
- |
- |
PREDICTED: short-chain type dehydrogenase/reductase isoform X2 [Jatropha curcas] |
| 7 |
Hb_003175_040 |
0.0770674677 |
- |
- |
PREDICTED: katanin p60 ATPase-containing subunit A1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_100506_010 |
0.0777269095 |
- |
- |
PREDICTED: uncharacterized protein LOC105133651 isoform X2 [Populus euphratica] |
| 9 |
Hb_003226_280 |
0.0793759731 |
- |
- |
PREDICTED: peter Pan-like protein [Populus euphratica] |
| 10 |
Hb_000865_070 |
0.0813152974 |
- |
- |
Protein FAR1-RELATED SEQUENCE 3 [Glycine soja] |
| 11 |
Hb_002527_070 |
0.0838889319 |
- |
- |
axonemal dynein light chain, putative [Ricinus communis] |
| 12 |
Hb_071736_020 |
0.0850052508 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 13 |
Hb_000261_460 |
0.0851148563 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 14 |
Hb_000671_150 |
0.0870356752 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 15 |
Hb_002028_130 |
0.0881738346 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 16 |
Hb_021650_030 |
0.0887002817 |
- |
- |
PREDICTED: uncharacterized protein LOC105642056 [Jatropha curcas] |
| 17 |
Hb_002078_450 |
0.0891024264 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 18 |
Hb_003408_020 |
0.0891313297 |
- |
- |
hypothetical protein RCOM_0453040 [Ricinus communis] |
| 19 |
Hb_001881_020 |
0.0913992936 |
- |
- |
clathrin binding protein, putative [Ricinus communis] |
| 20 |
Hb_000753_130 |
0.0915045616 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |