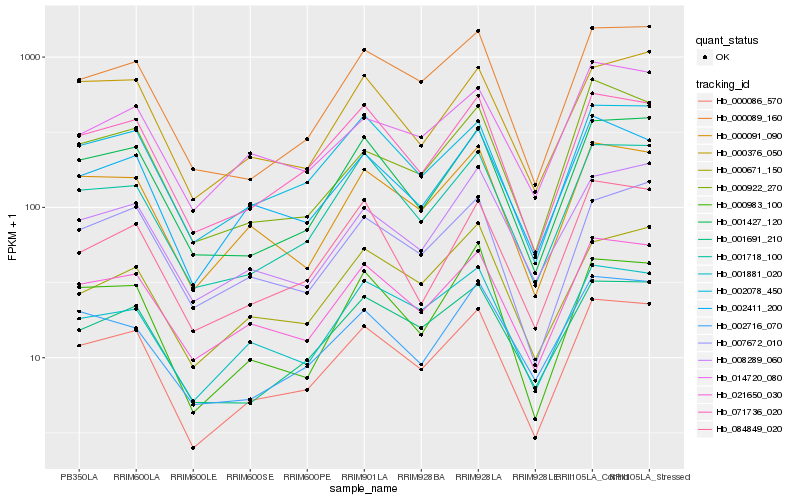
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_570 |
0.0 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 2 |
Hb_002078_450 |
0.0587357179 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 3 |
Hb_071736_020 |
0.0588398176 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 4 |
Hb_021650_030 |
0.0591683073 |
- |
- |
PREDICTED: uncharacterized protein LOC105642056 [Jatropha curcas] |
| 5 |
Hb_001427_120 |
0.0615228424 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 6 |
Hb_001718_100 |
0.0617318405 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 7 |
Hb_002411_200 |
0.0649284527 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000091_090 |
0.0653705309 |
- |
- |
hypothetical protein JCGZ_09031 [Jatropha curcas] |
| 9 |
Hb_000089_160 |
0.0667693573 |
- |
- |
60S ribosomal protein L5B [Hevea brasiliensis] |
| 10 |
Hb_001691_210 |
0.0740285377 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |
| 11 |
Hb_000376_050 |
0.0793076417 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 12 |
Hb_007672_010 |
0.0794983395 |
- |
- |
PREDICTED: uncharacterized protein LOC105639669 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001881_020 |
0.0816624548 |
- |
- |
clathrin binding protein, putative [Ricinus communis] |
| 14 |
Hb_000983_100 |
0.0816635595 |
- |
- |
PREDICTED: AP-4 complex subunit mu isoform X1 [Vitis vinifera] |
| 15 |
Hb_000922_270 |
0.0839808713 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 16 |
Hb_008289_060 |
0.084435302 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_084849_020 |
0.0847606936 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 18 |
Hb_014720_080 |
0.0848680522 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 19 |
Hb_000671_150 |
0.0855171826 |
- |
- |
PREDICTED: signal recognition particle 19 kDa protein-like [Jatropha curcas] |
| 20 |
Hb_002716_070 |
0.0875604269 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |