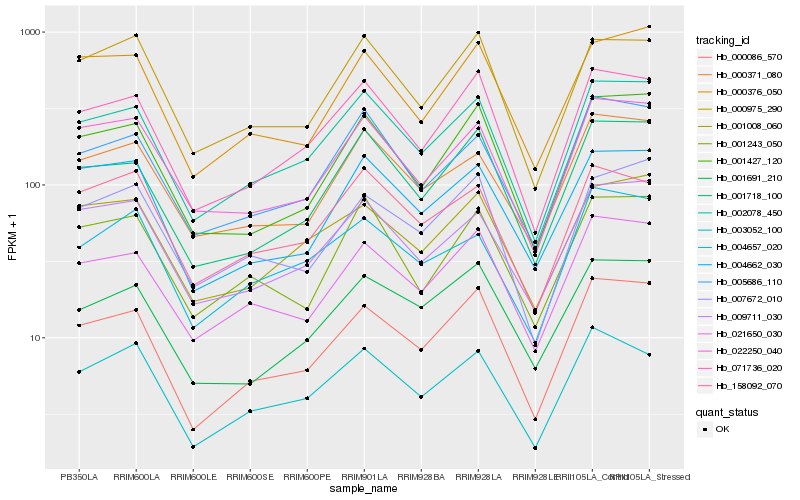
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002078_450 |
0.0 |
- |
- |
nuclear transport factor, putative [Ricinus communis] |
| 2 |
Hb_071736_020 |
0.047276904 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 3 |
Hb_005686_110 |
0.0558277918 |
- |
- |
PREDICTED: putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 2 [Populus euphratica] |
| 4 |
Hb_000086_570 |
0.0587357179 |
- |
- |
hypothetical protein JCGZ_24934 [Jatropha curcas] |
| 5 |
Hb_009711_030 |
0.0597252438 |
- |
- |
PREDICTED: uncharacterized protein LOC105647792 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001718_100 |
0.0604036701 |
- |
- |
PREDICTED: casein kinase II subunit beta-like isoform X5 [Jatropha curcas] |
| 7 |
Hb_022250_040 |
0.0607730358 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 2, mitochondrial [Jatropha curcas] |
| 8 |
Hb_021650_030 |
0.0621804264 |
- |
- |
PREDICTED: uncharacterized protein LOC105642056 [Jatropha curcas] |
| 9 |
Hb_000371_080 |
0.0651273908 |
- |
- |
ubiquitin-conjugating enzyme m, putative [Ricinus communis] |
| 10 |
Hb_001008_060 |
0.0659111662 |
- |
- |
PREDICTED: monothiol glutaredoxin-S15, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001427_120 |
0.0683512348 |
- |
- |
PREDICTED: uncharacterized protein LOC105637787 [Jatropha curcas] |
| 12 |
Hb_003052_100 |
0.0718300545 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_004662_030 |
0.073509657 |
- |
- |
hypothetical protein PRUPE_ppa012487mg [Prunus persica] |
| 14 |
Hb_158092_070 |
0.0737714685 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit Rieske-4, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_000975_290 |
0.0749068526 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 16 |
Hb_004657_020 |
0.0763352742 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 17 |
Hb_000376_050 |
0.0765555335 |
- |
- |
polyadenylate-binding protein, putative [Ricinus communis] |
| 18 |
Hb_001243_050 |
0.0775375741 |
- |
- |
PREDICTED: uncharacterized protein LOC105643776 [Jatropha curcas] |
| 19 |
Hb_001691_210 |
0.0778125775 |
transcription factor |
TF Family: C2C2-LSD |
hypothetical protein JCGZ_21436 [Jatropha curcas] |
| 20 |
Hb_007672_010 |
0.0785584219 |
- |
- |
PREDICTED: uncharacterized protein LOC105639669 isoform X1 [Jatropha curcas] |