| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
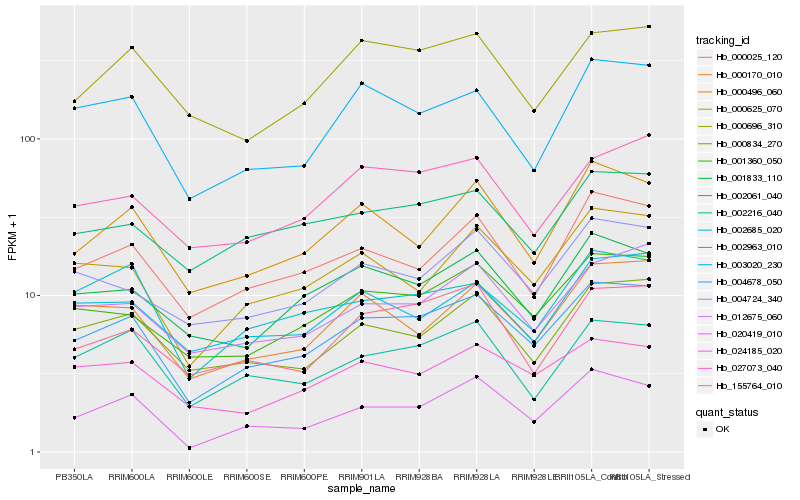
Hb_004678_050 |
0.0 |
- |
- |
PREDICTED: probable tRNA N6-adenosine threonylcarbamoyltransferase [Jatropha curcas] |
| 2 |
Hb_001360_050 |
0.0711473689 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000025_120 |
0.0854190347 |
- |
- |
PREDICTED: uncharacterized protein LOC105628946 isoform X1 [Jatropha curcas] |
| 4 |
Hb_012675_060 |
0.0857209405 |
- |
- |
hypothetical protein PRUPE_ppa007715mg [Prunus persica] |
| 5 |
Hb_000834_270 |
0.0857342789 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X2 [Jatropha curcas] |
| 6 |
Hb_020419_010 |
0.0873208682 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 7 |
Hb_003020_230 |
0.0879240414 |
- |
- |
DNA-directed RNA polymerase II subunit RPB7 [Glycine max] |
| 8 |
Hb_024185_020 |
0.0882463651 |
- |
- |
- |
| 9 |
Hb_000625_070 |
0.0885580201 |
transcription factor |
TF Family: CSD |
PREDICTED: cold shock domain-containing protein 3 [Jatropha curcas] |
| 10 |
Hb_001833_110 |
0.0895961732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_027073_040 |
0.091236281 |
- |
- |
TGF-beta-inducible nuclear protein, putative [Ricinus communis] |
| 12 |
Hb_000496_060 |
0.0912790302 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH3 [Jatropha curcas] |
| 13 |
Hb_002216_040 |
0.0924417919 |
- |
- |
PREDICTED: protein PRD1 [Jatropha curcas] |
| 14 |
Hb_004724_340 |
0.0925415314 |
- |
- |
PREDICTED: mitochondrial fission protein ELM1 [Jatropha curcas] |
| 15 |
Hb_000170_010 |
0.0933287903 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_155764_010 |
0.0937038644 |
- |
- |
- |
| 17 |
Hb_002061_040 |
0.0941281128 |
- |
- |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 18 |
Hb_002685_020 |
0.095247136 |
- |
- |
- |
| 19 |
Hb_000696_310 |
0.0954758884 |
- |
- |
PREDICTED: 40S ribosomal protein S4-3 [Jatropha curcas] |
| 20 |
Hb_002963_010 |
0.0957719106 |
- |
- |
PREDICTED: autophagy-related protein 18b-like isoform X1 [Jatropha curcas] |