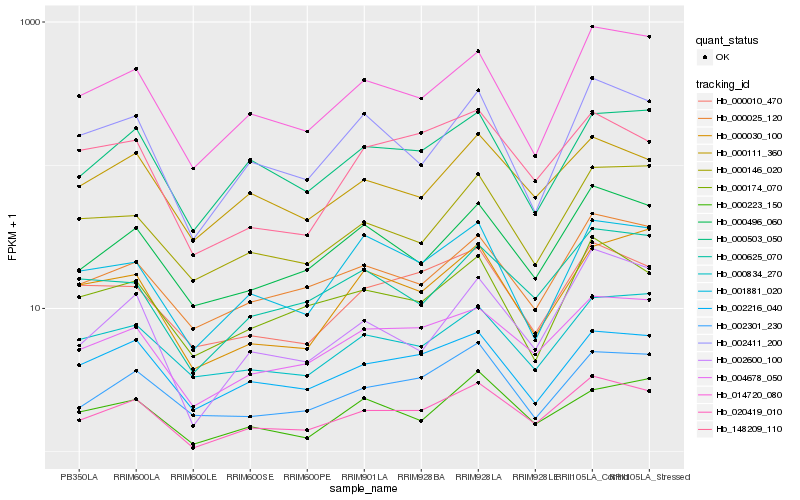
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020419_010 |
0.0 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |
| 2 |
Hb_004678_050 |
0.0873208682 |
- |
- |
PREDICTED: probable tRNA N6-adenosine threonylcarbamoyltransferase [Jatropha curcas] |
| 3 |
Hb_002600_100 |
0.1000184916 |
- |
- |
PREDICTED: putative endo-1,3(4)-beta-glucanase 2 [Jatropha curcas] |
| 4 |
Hb_014720_080 |
0.1041571151 |
- |
- |
iron-sulfur cluster scaffold protein [Hevea brasiliensis] |
| 5 |
Hb_000496_060 |
0.1052134525 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH3 [Jatropha curcas] |
| 6 |
Hb_002411_200 |
0.1107494015 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000025_120 |
0.1125712754 |
- |
- |
PREDICTED: uncharacterized protein LOC105628946 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000625_070 |
0.1127030786 |
transcription factor |
TF Family: CSD |
PREDICTED: cold shock domain-containing protein 3 [Jatropha curcas] |
| 9 |
Hb_002216_040 |
0.1143751532 |
- |
- |
PREDICTED: protein PRD1 [Jatropha curcas] |
| 10 |
Hb_000503_050 |
0.1159560078 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 1-like [Gossypium raimondii] |
| 11 |
Hb_002301_230 |
0.1176361635 |
- |
- |
hypothetical protein JCGZ_21466 [Jatropha curcas] |
| 12 |
Hb_148209_110 |
0.1191197919 |
- |
- |
putative polyol transported protein 2 [Hevea brasiliensis] |
| 13 |
Hb_000010_470 |
0.1191670546 |
- |
- |
Galactose oxidase/kelch repeat superfamily protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_000111_360 |
0.1224205391 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000223_150 |
0.1233292675 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49170, chloroplastic [Populus euphratica] |
| 16 |
Hb_000174_070 |
0.1234927876 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_001881_020 |
0.1241003251 |
- |
- |
clathrin binding protein, putative [Ricinus communis] |
| 18 |
Hb_000146_020 |
0.1241358286 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000834_270 |
0.1243982597 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000030_100 |
0.1244112737 |
- |
- |
PREDICTED: short-chain type dehydrogenase/reductase isoform X2 [Jatropha curcas] |