| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
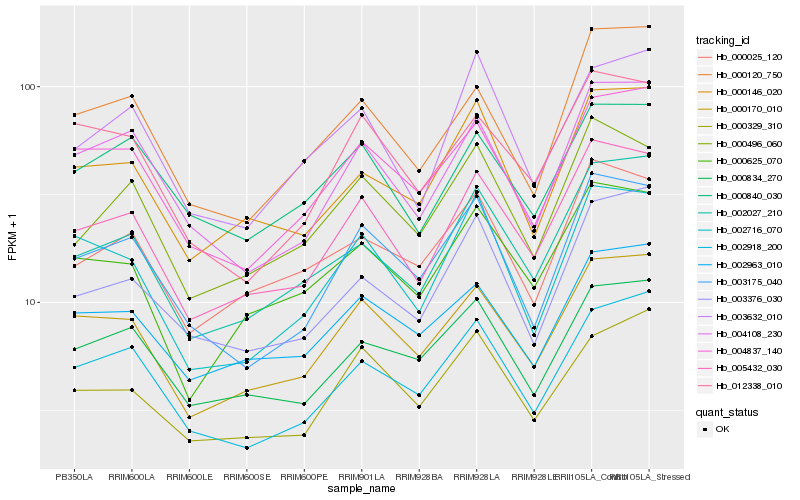
Hb_005432_030 |
0.0 |
- |
- |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 7 [Gossypium raimondii] |
| 2 |
Hb_000170_010 |
0.0524520118 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_002027_210 |
0.0605919275 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 4 |
Hb_000625_070 |
0.0643823648 |
transcription factor |
TF Family: CSD |
PREDICTED: cold shock domain-containing protein 3 [Jatropha curcas] |
| 5 |
Hb_000496_060 |
0.0681592552 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH3 [Jatropha curcas] |
| 6 |
Hb_004108_230 |
0.0728478477 |
- |
- |
PREDICTED: 6.7 kDa chloroplast outer envelope membrane protein-like [Jatropha curcas] |
| 7 |
Hb_003175_040 |
0.0752792947 |
- |
- |
PREDICTED: katanin p60 ATPase-containing subunit A1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000025_120 |
0.0756399478 |
- |
- |
PREDICTED: uncharacterized protein LOC105628946 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000146_020 |
0.0761066818 |
- |
- |
PREDICTED: protein TIC 21, chloroplastic [Jatropha curcas] |
| 10 |
Hb_004837_140 |
0.0768740629 |
- |
- |
hypothetical protein PRUPE_ppa012469mg [Prunus persica] |
| 11 |
Hb_012338_010 |
0.0779928937 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002963_010 |
0.0786590026 |
- |
- |
PREDICTED: autophagy-related protein 18b-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_002918_200 |
0.0793204519 |
- |
- |
- |
| 14 |
Hb_000834_270 |
0.079676117 |
- |
- |
PREDICTED: uncharacterized protein LOC105647032 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000120_750 |
0.0806174219 |
desease resistance |
Gene Name: NTPase_1 |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_002716_070 |
0.0817626459 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 17 |
Hb_000840_030 |
0.0836772146 |
transcription factor |
TF Family: IWS1 |
PREDICTED: probable mediator of RNA polymerase II transcription subunit 26c isoform X2 [Jatropha curcas] |
| 18 |
Hb_000329_310 |
0.0855838615 |
- |
- |
PREDICTED: phosphatidate cytidylyltransferase 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003376_030 |
0.0869641871 |
- |
- |
PREDICTED: ribonuclease P protein subunit p29 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003632_010 |
0.0872026938 |
- |
- |
conserved hypothetical protein [Ricinus communis] |