| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
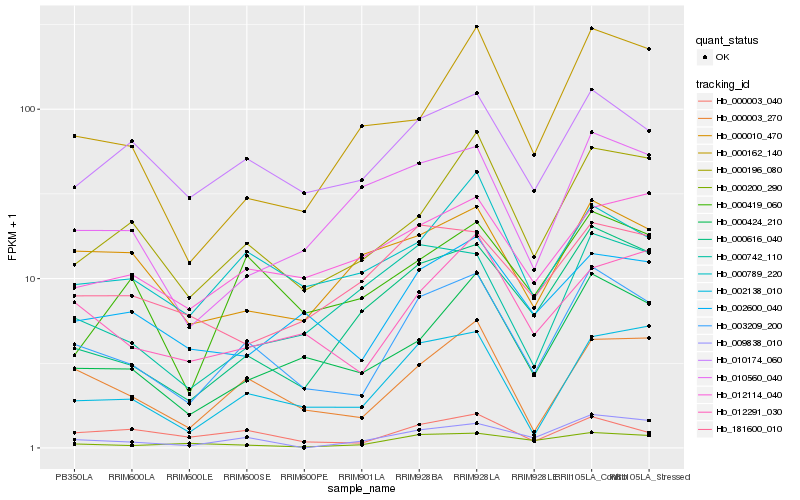
Hb_003209_200 |
0.0 |
transcription factor |
TF Family: WRKY |
hypothetical protein RCOM_1621230 [Ricinus communis] |
| 2 |
Hb_000003_270 |
0.1257067897 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 3 |
Hb_000196_080 |
0.1407806626 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000424_210 |
0.1418862784 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002138_010 |
0.144025735 |
- |
- |
hypothetical protein POPTR_0001s30620g [Populus trichocarpa] |
| 6 |
Hb_010174_060 |
0.1470192616 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 7 |
Hb_012291_030 |
0.1530044995 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X3 [Populus euphratica] |
| 8 |
Hb_002600_040 |
0.1534178166 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 9 |
Hb_000789_220 |
0.1536094325 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 10 |
Hb_000003_040 |
0.1537857953 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 11 |
Hb_000616_040 |
0.1586784467 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 12 |
Hb_009838_010 |
0.1604512033 |
- |
- |
- |
| 13 |
Hb_000200_290 |
0.1697619517 |
- |
- |
PREDICTED: elongator complex protein 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000419_060 |
0.1712276792 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |
| 15 |
Hb_012114_040 |
0.171389067 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000742_110 |
0.1714302871 |
- |
- |
hypothetical protein JCGZ_09215 [Jatropha curcas] |
| 17 |
Hb_010560_040 |
0.1744359866 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 11 [Jatropha curcas] |
| 18 |
Hb_000010_470 |
0.1745787964 |
- |
- |
Galactose oxidase/kelch repeat superfamily protein isoform 2 [Theobroma cacao] |
| 19 |
Hb_181600_010 |
0.178912927 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 20 |
Hb_000162_140 |
0.1791442852 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2 [Jatropha curcas] |