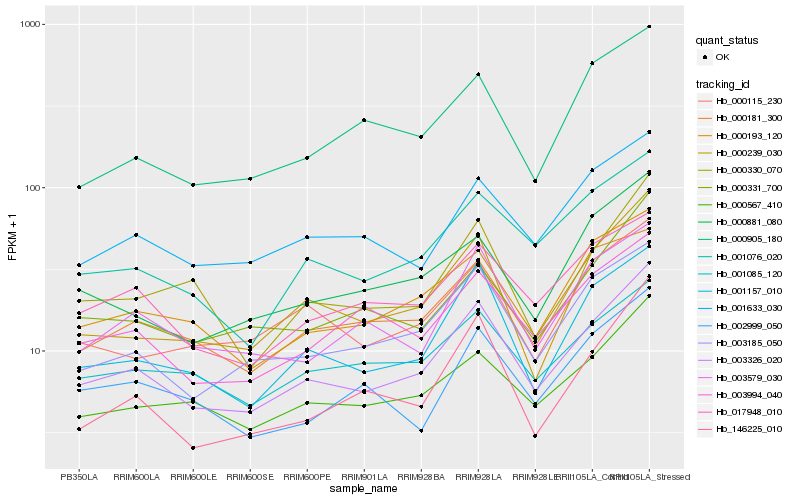
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003326_020 |
0.0 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 2 |
Hb_003185_050 |
0.0792066566 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 3 |
Hb_001157_010 |
0.0793257055 |
- |
- |
deoxyhypusine synthase, putative [Ricinus communis] |
| 4 |
Hb_000567_410 |
0.0821536818 |
- |
- |
hypothetical protein JCGZ_12784 [Jatropha curcas] |
| 5 |
Hb_001076_020 |
0.0846228229 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 8 [Eucalyptus grandis] |
| 6 |
Hb_000193_120 |
0.0847217359 |
- |
- |
PREDICTED: protein preY, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000239_030 |
0.085872452 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin 2-like [Jatropha curcas] |
| 8 |
Hb_001633_030 |
0.0908572456 |
- |
- |
- |
| 9 |
Hb_003579_030 |
0.0934657741 |
- |
- |
phosphatase 2C family protein [Populus trichocarpa] |
| 10 |
Hb_017948_010 |
0.0940776953 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |
| 11 |
Hb_000331_700 |
0.0950476361 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |
| 12 |
Hb_000330_070 |
0.0952273346 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 13 |
Hb_000881_080 |
0.096146363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000115_230 |
0.0970987508 |
- |
- |
PREDICTED: cysteine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 15 |
Hb_146225_010 |
0.0980273398 |
- |
- |
mitochondrial inner membrane protease subunit, putative [Ricinus communis] |
| 16 |
Hb_000181_300 |
0.0986846002 |
- |
- |
PREDICTED: uncharacterized protein LOC105650445 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001085_120 |
0.098854286 |
- |
- |
PREDICTED: UV radiation resistance-associated gene protein isoform X1 [Jatropha curcas] |
| 18 |
Hb_003994_040 |
0.1000081182 |
- |
- |
- |
| 19 |
Hb_002999_050 |
0.1027176611 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_000905_180 |
0.1035038863 |
- |
- |
60S ribosomal protein L31A [Hevea brasiliensis] |