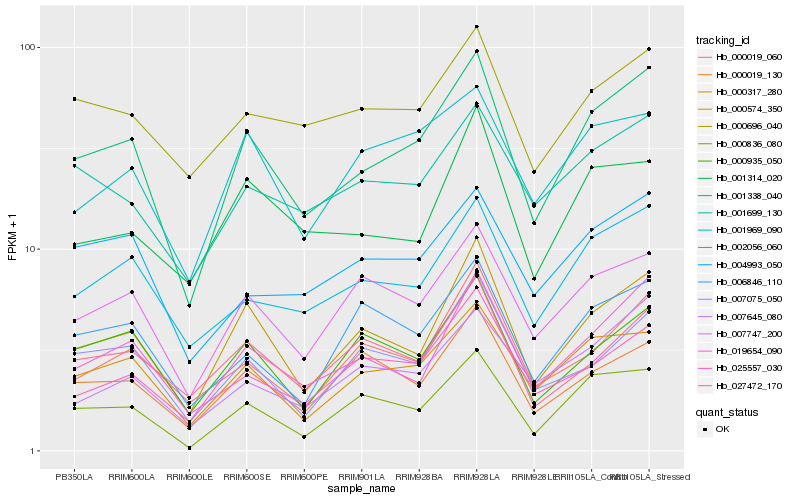
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001338_040 |
0.0 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_000317_280 |
0.0854118937 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative pentatricopeptide repeat-containing protein At5g08490 [Jatropha curcas] |
| 3 |
Hb_000574_350 |
0.0877470679 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 4 |
Hb_002056_060 |
0.0997513411 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_007747_200 |
0.1021297476 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g37570 [Jatropha curcas] |
| 6 |
Hb_000836_080 |
0.1048954725 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g46050, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000935_050 |
0.1157392791 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g49710 [Jatropha curcas] |
| 8 |
Hb_001699_130 |
0.1158478423 |
transcription factor |
TF Family: CSD |
PREDICTED: cold shock domain-containing protein 3 [Jatropha curcas] |
| 9 |
Hb_019654_090 |
0.1159762772 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33680 [Jatropha curcas] |
| 10 |
Hb_004993_050 |
0.1167793489 |
- |
- |
- |
| 11 |
Hb_001969_090 |
0.1189485966 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 12 |
Hb_007075_050 |
0.1194153093 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_025557_030 |
0.1196828611 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74600, chloroplastic [Jatropha curcas] |
| 14 |
Hb_007645_080 |
0.1207110079 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 15 |
Hb_000019_060 |
0.1222400805 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26782, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000696_040 |
0.1227547087 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 17 |
Hb_000019_130 |
0.1238796468 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At2g01510 [Jatropha curcas] |
| 18 |
Hb_006846_110 |
0.124081453 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61800 [Jatropha curcas] |
| 19 |
Hb_027472_170 |
0.1263522357 |
- |
- |
hypothetical protein POPTR_0007s08080g [Populus trichocarpa] |
| 20 |
Hb_001314_020 |
0.1280118671 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |