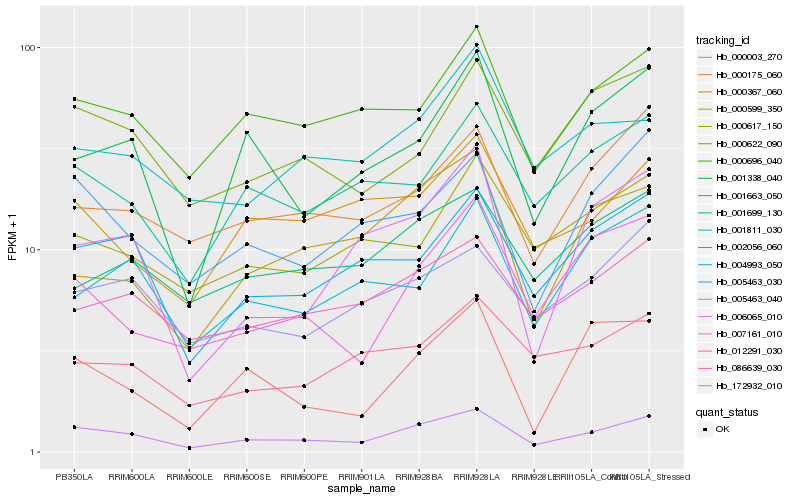
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006065_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639733 [Jatropha curcas] |
| 2 |
Hb_004993_050 |
0.1226818214 |
- |
- |
- |
| 3 |
Hb_005463_030 |
0.1257852872 |
- |
- |
PREDICTED: uncharacterized protein LOC105632512 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000696_040 |
0.1261380702 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 5 |
Hb_001338_040 |
0.1294707733 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 6 |
Hb_000622_090 |
0.1316371335 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 7 |
Hb_001699_130 |
0.1339114292 |
transcription factor |
TF Family: CSD |
PREDICTED: cold shock domain-containing protein 3 [Jatropha curcas] |
| 8 |
Hb_012291_030 |
0.1354560411 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X3 [Populus euphratica] |
| 9 |
Hb_086639_030 |
0.1359500986 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 10 |
Hb_000175_060 |
0.1382151634 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 11 |
Hb_005463_040 |
0.1399246648 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 12 |
Hb_007161_010 |
0.1406372565 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105630404 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000003_270 |
0.1433299927 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 14 |
Hb_000367_060 |
0.1455667485 |
- |
- |
diacylglycerol kinase, theta, putative [Ricinus communis] |
| 15 |
Hb_000599_350 |
0.1488117965 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_002056_060 |
0.1493246362 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001663_050 |
0.149378223 |
- |
- |
PREDICTED: GPN-loop GTPase 3 [Jatropha curcas] |
| 18 |
Hb_172932_010 |
0.1505773433 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_001811_030 |
0.1512026126 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 20 |
Hb_000617_150 |
0.1514535629 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61360-like [Jatropha curcas] |