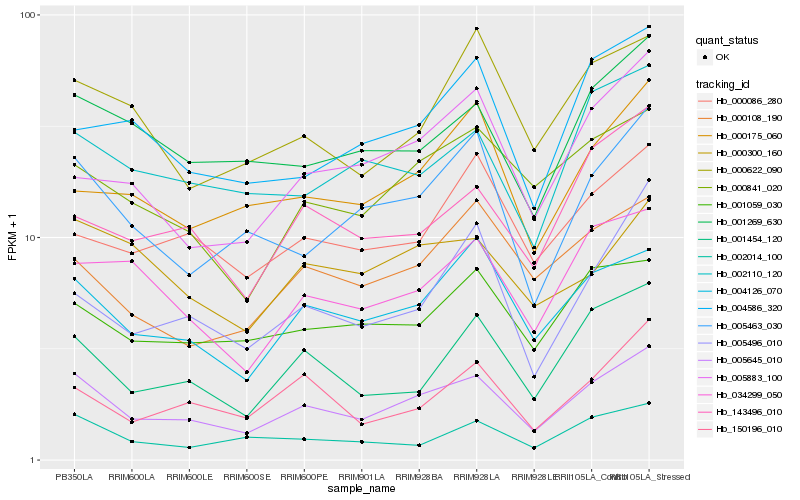
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005645_010 |
0.0 |
- |
- |
rhythmically-expressed protein 2 protein, putative [Ricinus communis] |
| 2 |
Hb_004126_070 |
0.0977268488 |
desease resistance |
Gene Name: CDC48_2 |
PREDICTED: vesicle-fusing ATPase [Jatropha curcas] |
| 3 |
Hb_005463_030 |
0.1079271383 |
- |
- |
PREDICTED: uncharacterized protein LOC105632512 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001454_120 |
0.1098012394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001269_630 |
0.110273207 |
- |
- |
PREDICTED: uncharacterized protein LOC105630301 [Jatropha curcas] |
| 6 |
Hb_000841_020 |
0.1124148974 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 7 |
Hb_034299_050 |
0.1149419558 |
- |
- |
hypothetical protein CICLE_v10031248mg [Citrus clementina] |
| 8 |
Hb_001059_030 |
0.1188869183 |
- |
- |
PREDICTED: F-box protein At5g03100-like [Solanum lycopersicum] |
| 9 |
Hb_002110_120 |
0.1195894187 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |
| 10 |
Hb_002014_100 |
0.1226122876 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_000108_190 |
0.1237793504 |
- |
- |
PREDICTED: dihydroorotate dehydrogenase (quinone), mitochondrial [Jatropha curcas] |
| 12 |
Hb_150196_010 |
0.1257474949 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 13 |
Hb_143496_010 |
0.1259363924 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3 [Glycine max] |
| 14 |
Hb_000622_090 |
0.1260260804 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 15 |
Hb_000175_060 |
0.1266178877 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 16 |
Hb_000086_280 |
0.1285756204 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 3-like [Gossypium raimondii] |
| 17 |
Hb_005496_010 |
0.1287179171 |
- |
- |
PREDICTED: PXMP2/4 family protein 4 [Jatropha curcas] |
| 18 |
Hb_000300_160 |
0.1290814349 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: cyclic dof factor 3-like [Jatropha curcas] |
| 19 |
Hb_005883_100 |
0.1295674057 |
- |
- |
5-formyltetrahydrofolate cyclo-ligase, putative [Ricinus communis] |
| 20 |
Hb_004586_320 |
0.1303927487 |
- |
- |
PREDICTED: bet1-like protein At4g14600 [Jatropha curcas] |