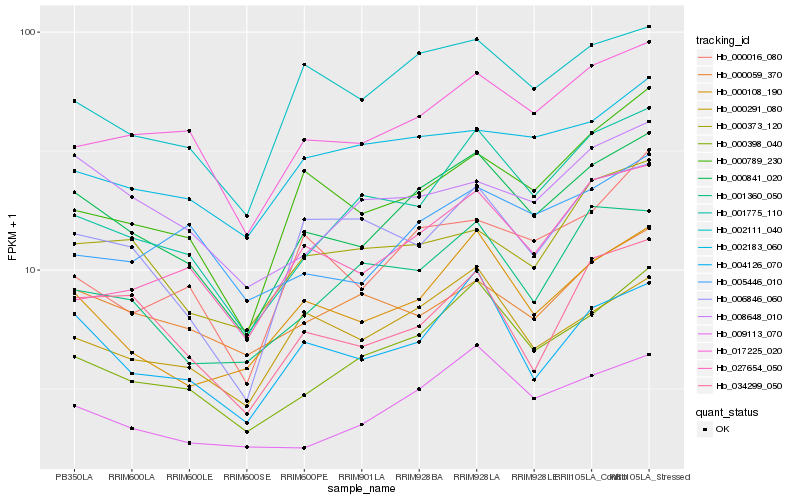
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000841_020 |
0.0 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 2 |
Hb_000398_040 |
0.0787296481 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 3 |
Hb_004126_070 |
0.0823615875 |
desease resistance |
Gene Name: CDC48_2 |
PREDICTED: vesicle-fusing ATPase [Jatropha curcas] |
| 4 |
Hb_017225_020 |
0.0841998593 |
- |
- |
PREDICTED: protein BRICK 1 [Gossypium raimondii] |
| 5 |
Hb_000108_190 |
0.0859645793 |
- |
- |
PREDICTED: dihydroorotate dehydrogenase (quinone), mitochondrial [Jatropha curcas] |
| 6 |
Hb_002111_040 |
0.0875810371 |
- |
- |
hypothetical protein JCGZ_16989 [Jatropha curcas] |
| 7 |
Hb_009113_070 |
0.0892733022 |
- |
- |
hypothetical protein POPTR_0010s07510g [Populus trichocarpa] |
| 8 |
Hb_000291_080 |
0.0938136369 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 9 |
Hb_001775_110 |
0.096776601 |
- |
- |
PREDICTED: probable ribosome biogenesis protein RLP24 [Jatropha curcas] |
| 10 |
Hb_000789_230 |
0.0991119271 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 11 |
Hb_034299_050 |
0.1008069995 |
- |
- |
hypothetical protein CICLE_v10031248mg [Citrus clementina] |
| 12 |
Hb_000373_120 |
0.1047298318 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 13 |
Hb_002183_060 |
0.1048864164 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_008648_010 |
0.1050644123 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Jatropha curcas] |
| 15 |
Hb_006846_060 |
0.1062236271 |
- |
- |
inositol hexaphosphate kinase, putative [Ricinus communis] |
| 16 |
Hb_001360_050 |
0.1068939502 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_027654_050 |
0.1081881254 |
- |
- |
PREDICTED: uncharacterized protein LOC105646592 [Jatropha curcas] |
| 18 |
Hb_005446_010 |
0.1096292462 |
- |
- |
PREDICTED: zinc finger protein 830 [Jatropha curcas] |
| 19 |
Hb_000016_080 |
0.110039771 |
- |
- |
PREDICTED: uncharacterized protein LOC105648533 [Jatropha curcas] |
| 20 |
Hb_000059_370 |
0.1102781467 |
- |
- |
PREDICTED: U6 snRNA phosphodiesterase [Jatropha curcas] |