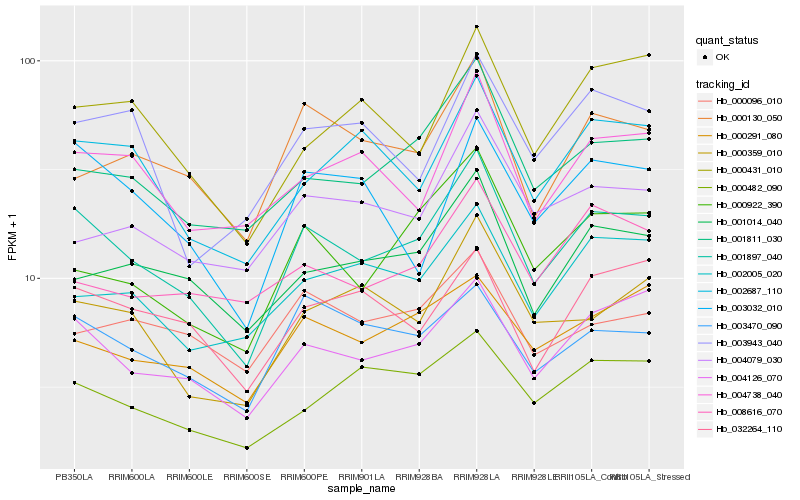
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001897_040 |
0.0 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 2 |
Hb_004126_070 |
0.0837496598 |
desease resistance |
Gene Name: CDC48_2 |
PREDICTED: vesicle-fusing ATPase [Jatropha curcas] |
| 3 |
Hb_001811_030 |
0.1022322761 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 4 |
Hb_000922_390 |
0.1100449539 |
- |
- |
PREDICTED: uncharacterized protein LOC105640364 [Jatropha curcas] |
| 5 |
Hb_001014_040 |
0.1121216399 |
- |
- |
PREDICTED: uncharacterized protein LOC105647308 [Jatropha curcas] |
| 6 |
Hb_003470_090 |
0.1121968461 |
- |
- |
PREDICTED: pumilio homolog 12 [Jatropha curcas] |
| 7 |
Hb_000291_080 |
0.1147803169 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 8 |
Hb_002687_110 |
0.1156708493 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C343.04c isoform X1 [Jatropha curcas] |
| 9 |
Hb_004738_040 |
0.1160421033 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1-like isoform X2 [Populus euphratica] |
| 10 |
Hb_000130_050 |
0.1186008889 |
- |
- |
14-3-3 protein, putative [Ricinus communis] |
| 11 |
Hb_003032_010 |
0.1190168872 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 12 |
Hb_000359_010 |
0.1192574976 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 13 |
Hb_000482_090 |
0.1193240592 |
- |
- |
PREDICTED: uncharacterized protein LOC105638145 [Jatropha curcas] |
| 14 |
Hb_032264_110 |
0.1203695824 |
- |
- |
PREDICTED: uncharacterized protein LOC105648219 [Jatropha curcas] |
| 15 |
Hb_002005_020 |
0.1203817394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004079_030 |
0.1263656103 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000431_010 |
0.1265840723 |
- |
- |
hypothetical protein POPTR_0016s05340g [Populus trichocarpa] |
| 18 |
Hb_003943_040 |
0.130423146 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_000096_010 |
0.1310342444 |
- |
- |
PREDICTED: U-box domain-containing protein 30-like [Citrus sinensis] |
| 20 |
Hb_008616_070 |
0.1310624702 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 8 isoform X1 [Jatropha curcas] |