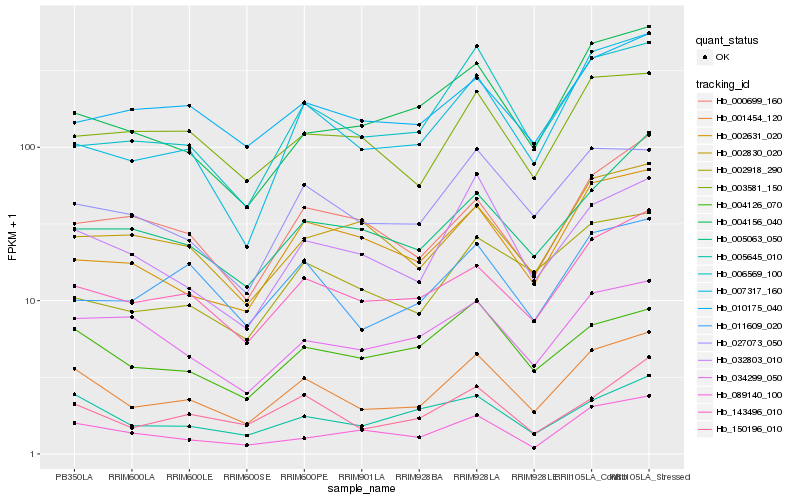
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_143496_010 |
0.0979217452 |
- |
- |
PREDICTED: 40S ribosomal protein S3-3 [Glycine max] |
| 3 |
Hb_150196_010 |
0.1053991891 |
- |
- |
PREDICTED: uncharacterized protein LOC105628218 isoform X1 [Jatropha curcas] |
| 4 |
Hb_032803_010 |
0.1072531925 |
- |
- |
PREDICTED: glycosyltransferase-like At2g41451 [Jatropha curcas] |
| 5 |
Hb_027073_050 |
0.1092037171 |
- |
- |
PREDICTED: uncharacterized protein LOC105635813 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005645_010 |
0.1098012394 |
- |
- |
rhythmically-expressed protein 2 protein, putative [Ricinus communis] |
| 7 |
Hb_007317_160 |
0.1199286521 |
- |
- |
PREDICTED: uncharacterized protein At2g27730, mitochondrial [Jatropha curcas] |
| 8 |
Hb_003581_150 |
0.1220927227 |
- |
- |
hypothetical protein CICLE_v10032366mg [Citrus clementina] |
| 9 |
Hb_011609_020 |
0.1226605308 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004126_070 |
0.1232333345 |
desease resistance |
Gene Name: CDC48_2 |
PREDICTED: vesicle-fusing ATPase [Jatropha curcas] |
| 11 |
Hb_010175_040 |
0.1249665197 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 6a, mitochondrial [Jatropha curcas] |
| 12 |
Hb_089140_100 |
0.1252939532 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4 [Jatropha curcas] |
| 13 |
Hb_004156_040 |
0.125602277 |
- |
- |
PREDICTED: uncharacterized protein LOC105110611 [Populus euphratica] |
| 14 |
Hb_002631_020 |
0.125694745 |
- |
- |
PREDICTED: protein FAM32A-like [Jatropha curcas] |
| 15 |
Hb_005063_050 |
0.1256957876 |
- |
- |
PREDICTED: probable prefoldin subunit 4 isoform X1 [Jatropha curcas] |
| 16 |
Hb_034299_050 |
0.1257141884 |
- |
- |
hypothetical protein CICLE_v10031248mg [Citrus clementina] |
| 17 |
Hb_006569_100 |
0.1257996714 |
- |
- |
hypothetical protein POPTR_0005s26990g [Populus trichocarpa] |
| 18 |
Hb_002918_290 |
0.1271034715 |
- |
- |
PREDICTED: uncharacterized protein LOC105649583 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002830_020 |
0.1276817495 |
- |
- |
PREDICTED: outer envelope pore protein 16-3, chloroplastic/mitochondrial [Jatropha curcas] |
| 20 |
Hb_000699_160 |
0.1278530134 |
- |
- |
PREDICTED: C-Myc-binding protein homolog [Jatropha curcas] |