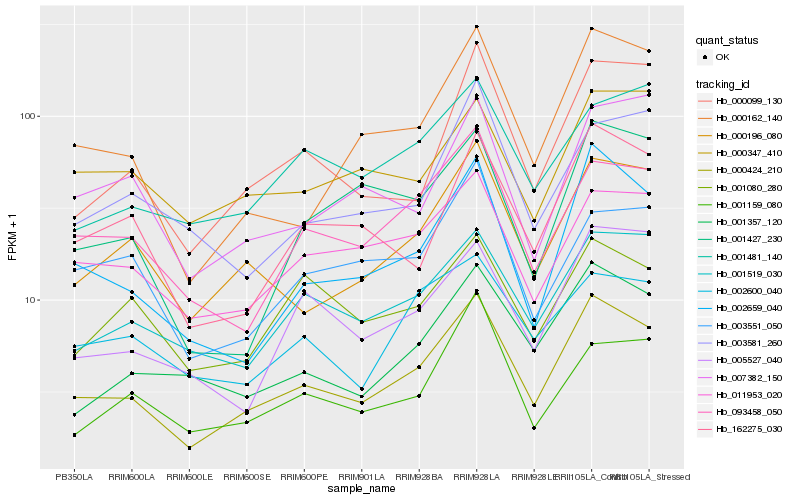
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000424_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002659_040 |
0.0941821708 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000196_080 |
0.1042417796 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH79-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_001519_030 |
0.1108190343 |
- |
- |
PREDICTED: protein trichome birefringence-like 12 isoform X1 [Jatropha curcas] |
| 5 |
Hb_093458_050 |
0.1112930759 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 6 |
Hb_000162_140 |
0.111589722 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2 [Jatropha curcas] |
| 7 |
Hb_000099_130 |
0.1130733886 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 8 |
Hb_162275_030 |
0.1141496862 |
- |
- |
PREDICTED: uncharacterized protein LOC105646638 [Jatropha curcas] |
| 9 |
Hb_011953_020 |
0.1167999586 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 10 |
Hb_001357_120 |
0.1184119111 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 11 |
Hb_005527_040 |
0.127305679 |
- |
- |
PREDICTED: methionine adenosyltransferase 2 subunit beta [Jatropha curcas] |
| 12 |
Hb_002600_040 |
0.1340378954 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 13 |
Hb_001427_230 |
0.1341550451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001481_140 |
0.1342353147 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 15 |
Hb_003551_050 |
0.1346992476 |
- |
- |
PREDICTED: mitochondrial fission 1 protein A [Jatropha curcas] |
| 16 |
Hb_001159_080 |
0.134936205 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 17 |
Hb_001080_280 |
0.1357831061 |
- |
- |
PREDICTED: uncharacterized protein LOC105649981 [Jatropha curcas] |
| 18 |
Hb_000347_410 |
0.1360429179 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 9 [Jatropha curcas] |
| 19 |
Hb_007382_150 |
0.1367773027 |
- |
- |
PREDICTED: GDT1-like protein 4 [Jatropha curcas] |
| 20 |
Hb_003581_260 |
0.1370496305 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |