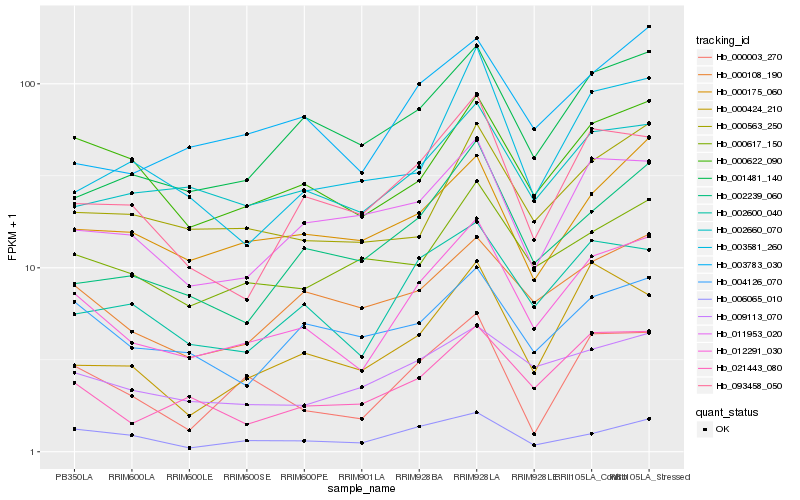
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012291_030 |
0.0 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN2 isoform X3 [Populus euphratica] |
| 2 |
Hb_002600_040 |
0.105659339 |
- |
- |
PREDICTED: probable transmembrane ascorbate ferrireductase 2 [Jatropha curcas] |
| 3 |
Hb_021443_080 |
0.1133510336 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 4 |
Hb_000622_090 |
0.1197680814 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 5 |
Hb_093458_050 |
0.1218230013 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 6 |
Hb_003783_030 |
0.1228605599 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |
| 7 |
Hb_000108_190 |
0.1260512221 |
- |
- |
PREDICTED: dihydroorotate dehydrogenase (quinone), mitochondrial [Jatropha curcas] |
| 8 |
Hb_002239_060 |
0.126763648 |
- |
- |
PREDICTED: general transcription factor IIH subunit 2 [Jatropha curcas] |
| 9 |
Hb_009113_070 |
0.1277152712 |
- |
- |
hypothetical protein POPTR_0010s07510g [Populus trichocarpa] |
| 10 |
Hb_000563_250 |
0.1304332858 |
- |
- |
PREDICTED: uncharacterized protein LOC105643472 [Jatropha curcas] |
| 11 |
Hb_002660_070 |
0.1328251138 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |
| 12 |
Hb_011953_020 |
0.1331510867 |
- |
- |
PREDICTED: ORM1-like protein 1 [Jatropha curcas] |
| 13 |
Hb_006065_010 |
0.1354560411 |
- |
- |
PREDICTED: uncharacterized protein LOC105639733 [Jatropha curcas] |
| 14 |
Hb_000003_270 |
0.1364107591 |
- |
- |
PREDICTED: cytokinin dehydrogenase 6 [Jatropha curcas] |
| 15 |
Hb_000175_060 |
0.1377556245 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 16 |
Hb_000617_150 |
0.1384596802 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61360-like [Jatropha curcas] |
| 17 |
Hb_004126_070 |
0.1400942583 |
desease resistance |
Gene Name: CDC48_2 |
PREDICTED: vesicle-fusing ATPase [Jatropha curcas] |
| 18 |
Hb_000424_210 |
0.1403897652 |
- |
- |
PREDICTED: uncharacterized protein LOC105638063 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001481_140 |
0.1403998971 |
- |
- |
PREDICTED: putative dual specificity protein phosphatase DSP8 [Jatropha curcas] |
| 20 |
Hb_003581_260 |
0.1405674038 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |