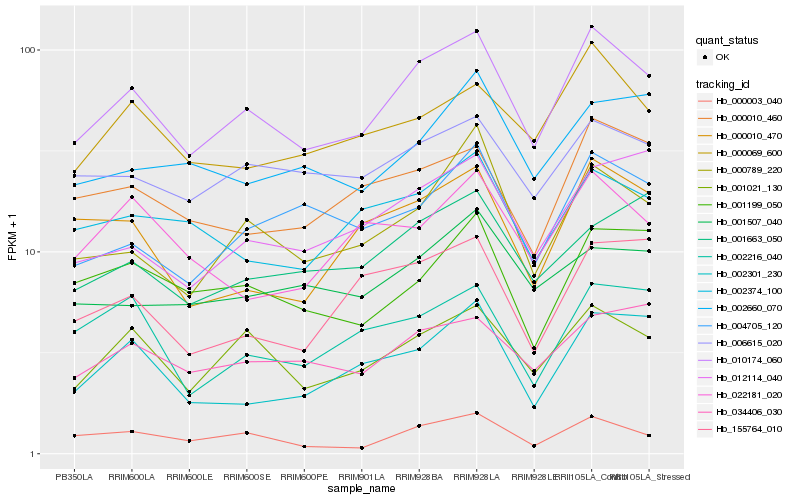
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010174_060 |
0.0 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 2 |
Hb_001021_130 |
0.081831245 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 3 |
Hb_000069_600 |
0.1058890121 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 4 |
Hb_006615_020 |
0.1077391478 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM2 [Jatropha curcas] |
| 5 |
Hb_000789_220 |
0.1096343019 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 6 |
Hb_000010_460 |
0.1111468649 |
- |
- |
hypothetical protein EUGRSUZ_B001502, partial [Eucalyptus grandis] |
| 7 |
Hb_034406_030 |
0.1117106673 |
- |
- |
PREDICTED: dihydroorotate dehydrogenase (quinone), mitochondrial [Musa acuminata subsp. malaccensis] |
| 8 |
Hb_000003_040 |
0.1125390624 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 9 |
Hb_002374_100 |
0.1142018067 |
- |
- |
replication factor C / DNA polymerase III gamma-tau subunit, putative [Ricinus communis] |
| 10 |
Hb_004705_120 |
0.1146890717 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_012114_040 |
0.1195699917 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_001199_050 |
0.1197625759 |
- |
- |
PREDICTED: DNA-3-methyladenine glycosylase-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000010_470 |
0.1202332133 |
- |
- |
Galactose oxidase/kelch repeat superfamily protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_001507_040 |
0.1247352497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002216_040 |
0.1249095935 |
- |
- |
PREDICTED: protein PRD1 [Jatropha curcas] |
| 16 |
Hb_155764_010 |
0.126137734 |
- |
- |
- |
| 17 |
Hb_001663_050 |
0.1262802551 |
- |
- |
PREDICTED: GPN-loop GTPase 3 [Jatropha curcas] |
| 18 |
Hb_002660_070 |
0.1264930412 |
- |
- |
PREDICTED: gamma carbonic anhydrase 1, mitochondrial [Eucalyptus grandis] |
| 19 |
Hb_002301_230 |
0.1265622894 |
- |
- |
hypothetical protein JCGZ_21466 [Jatropha curcas] |
| 20 |
Hb_022181_020 |
0.1268397297 |
- |
- |
PREDICTED: D-tyrosyl-tRNA(Tyr) deacylase isoform X1 [Jatropha curcas] |