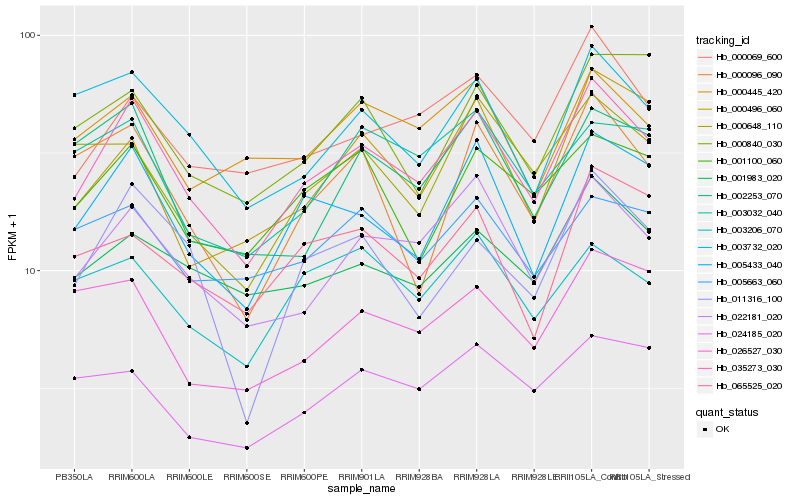
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_035273_030 |
0.0 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |
| 2 |
Hb_005433_040 |
0.0819655173 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 3 |
Hb_022181_020 |
0.0821574167 |
- |
- |
PREDICTED: D-tyrosyl-tRNA(Tyr) deacylase isoform X1 [Jatropha curcas] |
| 4 |
Hb_000069_600 |
0.097258392 |
- |
- |
PREDICTED: mitochondrial adenine nucleotide transporter ADNT1 [Jatropha curcas] |
| 5 |
Hb_001983_020 |
0.0995668411 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5 [Jatropha curcas] |
| 6 |
Hb_003732_020 |
0.1013859002 |
- |
- |
PREDICTED: phosphoglucomutase, chloroplastic-like [Populus euphratica] |
| 7 |
Hb_001100_060 |
0.1048422818 |
- |
- |
glycoprotein 3-alpha-l-fucosyltransferase A, putative [Ricinus communis] |
| 8 |
Hb_011316_100 |
0.1082299829 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4-like [Jatropha curcas] |
| 9 |
Hb_000445_420 |
0.1086095502 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 10 |
Hb_005663_060 |
0.1134873106 |
- |
- |
PREDICTED: uncharacterized protein LOC105770600 [Gossypium raimondii] |
| 11 |
Hb_002253_070 |
0.1137237732 |
transcription factor |
TF Family: NAC |
NAC transcription factor 042 [Jatropha curcas] |
| 12 |
Hb_003206_070 |
0.1144286286 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 13 |
Hb_065525_020 |
0.1149827059 |
- |
- |
PREDICTED: geranylgeranyl transferase type-1 subunit beta [Jatropha curcas] |
| 14 |
Hb_000840_030 |
0.1151787709 |
transcription factor |
TF Family: IWS1 |
PREDICTED: probable mediator of RNA polymerase II transcription subunit 26c isoform X2 [Jatropha curcas] |
| 15 |
Hb_000496_060 |
0.1153933507 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH3 [Jatropha curcas] |
| 16 |
Hb_003032_040 |
0.1154925345 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: zinc finger CCCH domain-containing protein 25 [Jatropha curcas] |
| 17 |
Hb_026527_030 |
0.1156866154 |
- |
- |
PREDICTED: general transcription factor IIE subunit 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000648_110 |
0.1164672588 |
- |
- |
PREDICTED: D-xylose-proton symporter-like 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_024185_020 |
0.1181246114 |
- |
- |
- |
| 20 |
Hb_000096_090 |
0.1186506219 |
- |
- |
hypothetical protein JCGZ_12551 [Jatropha curcas] |