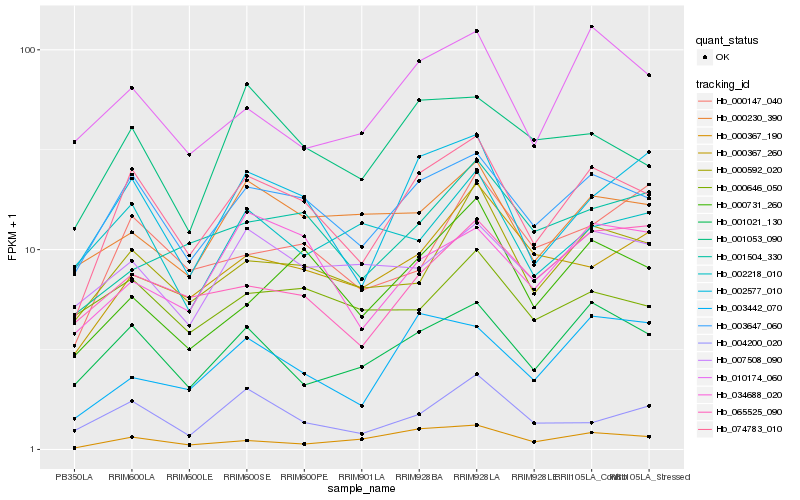
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_074783_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649970 [Jatropha curcas] |
| 2 |
Hb_003647_060 |
0.0678522475 |
- |
- |
PREDICTED: uncharacterized protein LOC105646679 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002577_010 |
0.1138163616 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 4 |
Hb_001021_130 |
0.1155375826 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 5 |
Hb_000592_020 |
0.1249213885 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 6 |
Hb_001053_090 |
0.1328994377 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 30-like [Jatropha curcas] |
| 7 |
Hb_034688_020 |
0.1421396288 |
- |
- |
PREDICTED: uncharacterized protein LOC105642050 [Jatropha curcas] |
| 8 |
Hb_000230_390 |
0.1423396953 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880 [Jatropha curcas] |
| 9 |
Hb_007508_090 |
0.1439273546 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 10 |
Hb_000731_260 |
0.1442357049 |
- |
- |
hypothetical protein RCOM_0905090 [Ricinus communis] |
| 11 |
Hb_010174_060 |
0.1442411522 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 12 |
Hb_000367_190 |
0.147243796 |
- |
- |
hypothetical protein RCOM_1470550 [Ricinus communis] |
| 13 |
Hb_000367_260 |
0.1485509023 |
- |
- |
PREDICTED: uncharacterized protein LOC105640585 [Jatropha curcas] |
| 14 |
Hb_003442_070 |
0.1509742282 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 15 |
Hb_004200_020 |
0.1534673482 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 16 |
Hb_000646_050 |
0.1558226572 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 22-like [Jatropha curcas] |
| 17 |
Hb_002218_010 |
0.156103763 |
transcription factor |
TF Family: Orphans |
ethylene receptor, putative [Ricinus communis] |
| 18 |
Hb_065525_090 |
0.1568397955 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 19 |
Hb_000147_040 |
0.1573910289 |
- |
- |
PREDICTED: uncharacterized protein LOC105638298 [Jatropha curcas] |
| 20 |
Hb_001504_330 |
0.1575197571 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |