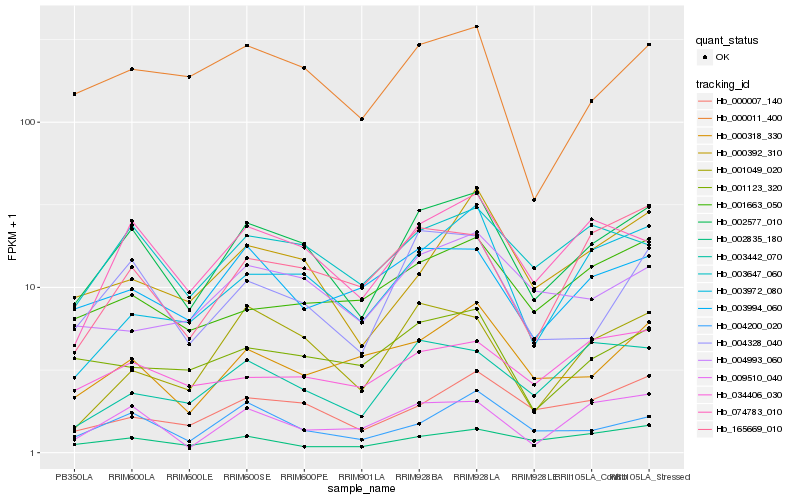
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002577_010 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 2 |
Hb_074783_010 |
0.1138163616 |
- |
- |
PREDICTED: uncharacterized protein LOC105649970 [Jatropha curcas] |
| 3 |
Hb_004328_040 |
0.1178036344 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 8 [Vitis vinifera] |
| 4 |
Hb_003647_060 |
0.1240210645 |
- |
- |
PREDICTED: uncharacterized protein LOC105646679 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000007_140 |
0.132020542 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 6 |
Hb_004200_020 |
0.1378902187 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 7 |
Hb_165669_010 |
0.1397394446 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_003994_060 |
0.1432356287 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000392_310 |
0.143729989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000011_400 |
0.1437732359 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 4 [Hevea brasiliensis] |
| 11 |
Hb_000318_330 |
0.1468402583 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g15630, mitochondrial [Jatropha curcas] |
| 12 |
Hb_001049_020 |
0.1478017957 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002835_180 |
0.1478407136 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_003442_070 |
0.1498100522 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 15 |
Hb_009510_040 |
0.1500745414 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_001123_320 |
0.1502310373 |
- |
- |
PREDICTED: uncharacterized protein LOC105647314 isoform X3 [Jatropha curcas] |
| 17 |
Hb_003972_080 |
0.1505599125 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 18 |
Hb_001663_050 |
0.1514209152 |
- |
- |
PREDICTED: GPN-loop GTPase 3 [Jatropha curcas] |
| 19 |
Hb_034406_030 |
0.1521249734 |
- |
- |
PREDICTED: dihydroorotate dehydrogenase (quinone), mitochondrial [Musa acuminata subsp. malaccensis] |
| 20 |
Hb_004993_060 |
0.1529004564 |
- |
- |
- |