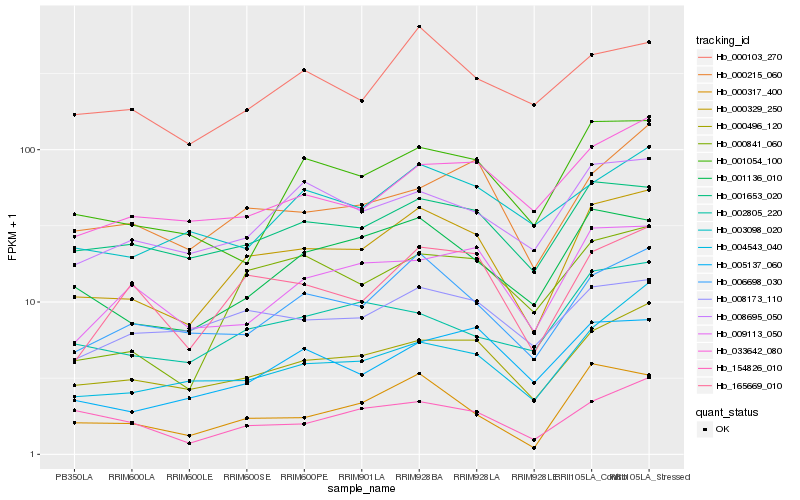
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_250 |
0.0 |
- |
- |
PREDICTED: vesicle-associated protein 1-3 isoform X1 [Fragaria vesca subsp. vesca] |
| 2 |
Hb_000496_120 |
0.0953823491 |
- |
- |
PREDICTED: iron-sulfur protein NUBPL [Jatropha curcas] |
| 3 |
Hb_006698_030 |
0.1078474128 |
- |
- |
PREDICTED: uncharacterized protein LOC105633969 [Jatropha curcas] |
| 4 |
Hb_001136_010 |
0.114460963 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 5 |
Hb_165669_010 |
0.1144786431 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000317_400 |
0.1194693451 |
transcription factor |
TF Family: zf-HD |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001653_020 |
0.1207917102 |
transcription factor |
TF Family: C2H2 |
rar1, putative [Ricinus communis] |
| 8 |
Hb_000841_060 |
0.1225560415 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0015s06470g [Populus trichocarpa] |
| 9 |
Hb_008695_050 |
0.1250089687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001054_100 |
0.131740193 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 11 |
Hb_000215_060 |
0.1318259427 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM17-2-like [Jatropha curcas] |
| 12 |
Hb_005137_060 |
0.132046082 |
- |
- |
- |
| 13 |
Hb_033642_080 |
0.1320832574 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 14 |
Hb_004543_040 |
0.1325803544 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 15 |
Hb_009113_050 |
0.1329198859 |
- |
- |
PREDICTED: uncharacterized protein LOC105644587 [Jatropha curcas] |
| 16 |
Hb_002805_220 |
0.136394261 |
- |
- |
PREDICTED: 60S ribosomal protein L2, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000103_270 |
0.1381597463 |
- |
- |
PREDICTED: 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase 2 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_008173_110 |
0.1387077429 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_154826_010 |
0.1399828771 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_003098_020 |
0.1404934993 |
- |
- |
40S ribosomal protein S5B [Hevea brasiliensis] |