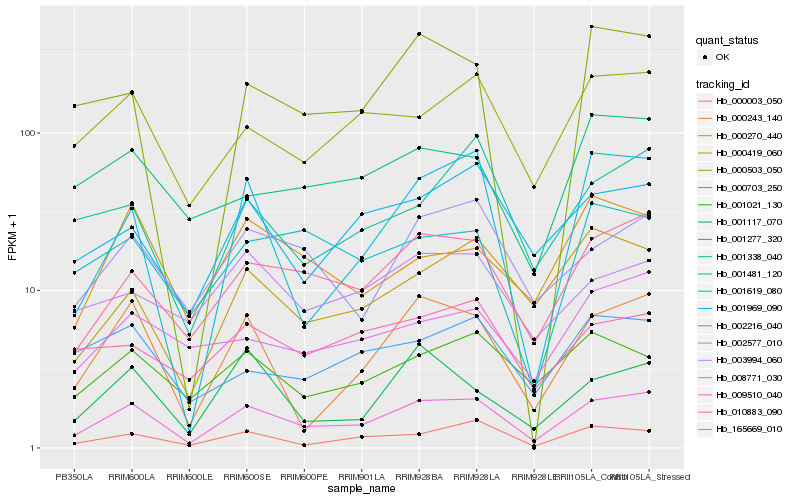
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009510_040 |
0.0 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen2-1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_165669_010 |
0.1107068957 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000243_140 |
0.138310012 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 4 |
Hb_000503_050 |
0.1417580053 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 1-like [Gossypium raimondii] |
| 5 |
Hb_000003_050 |
0.1462213477 |
- |
- |
Cysteine-rich RLK (RECEPTOR-like protein kinase) 8 [Theobroma cacao] |
| 6 |
Hb_002577_010 |
0.1500745414 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 7 |
Hb_000419_060 |
0.1541666238 |
- |
- |
PREDICTED: patatin-like protein 6 [Jatropha curcas] |
| 8 |
Hb_001969_090 |
0.1547773502 |
- |
- |
plastid ATP/ADP transport protein 2 [Manihot esculenta] |
| 9 |
Hb_001481_120 |
0.1630936617 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 10 |
Hb_001021_130 |
0.1634577465 |
- |
- |
PREDICTED: uncharacterized protein LOC105642479 [Jatropha curcas] |
| 11 |
Hb_001619_080 |
0.1643259732 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001338_040 |
0.1661687474 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_010883_090 |
0.167459266 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 14 |
Hb_008771_030 |
0.1676127665 |
- |
- |
PREDICTED: uncharacterized protein LOC105642119 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000270_440 |
0.1682957141 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0015s08130g [Populus trichocarpa] |
| 16 |
Hb_002216_040 |
0.1684679933 |
- |
- |
PREDICTED: protein PRD1 [Jatropha curcas] |
| 17 |
Hb_001117_070 |
0.1693798947 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 18 |
Hb_001277_320 |
0.1701832349 |
- |
- |
Stem-specific protein TSJT1, putative [Ricinus communis] |
| 19 |
Hb_003994_060 |
0.1713555082 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000703_250 |
0.1729011237 |
- |
- |
PREDICTED: protein LIGHT-DEPENDENT SHORT HYPOCOTYLS 10-like [Jatropha curcas] |