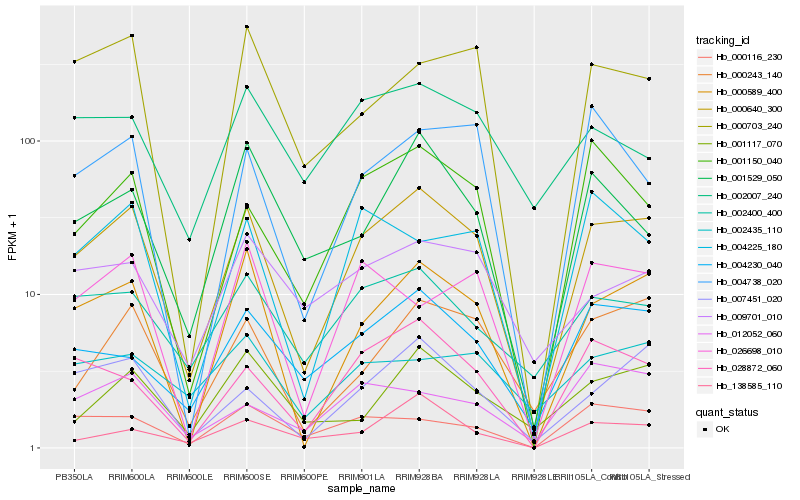
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000640_300 |
0.0 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 2 |
Hb_000589_400 |
0.1299526502 |
- |
- |
ESC, putative [Ricinus communis] |
| 3 |
Hb_000243_140 |
0.1587725814 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 4 |
Hb_007451_020 |
0.1591446202 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 5 |
Hb_028872_060 |
0.1636448142 |
- |
- |
PREDICTED: ankyrin-3-like [Jatropha curcas] |
| 6 |
Hb_001150_040 |
0.1651187785 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Solanum lycopersicum] |
| 7 |
Hb_004230_040 |
0.1661685778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_002400_400 |
0.1720564195 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 9 |
Hb_000703_240 |
0.1766398397 |
- |
- |
- |
| 10 |
Hb_000116_230 |
0.1774662848 |
- |
- |
- |
| 11 |
Hb_004225_180 |
0.1777939649 |
- |
- |
bel1 homeotic protein, putative [Ricinus communis] |
| 12 |
Hb_004738_020 |
0.1814170026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001529_050 |
0.1836795729 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 14 |
Hb_026698_010 |
0.1847481921 |
- |
- |
2,4-dienoyl-CoA reductase, putative [Ricinus communis] |
| 15 |
Hb_001117_070 |
0.1873116903 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 16 |
Hb_009701_010 |
0.1896105318 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 17 |
Hb_002435_110 |
0.1950206495 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromodomain-helicase-DNA-binding protein 1-like [Jatropha curcas] |
| 18 |
Hb_138585_110 |
0.1955961769 |
- |
- |
- |
| 19 |
Hb_002007_240 |
0.1975802941 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 20 |
Hb_012052_060 |
0.1976851505 |
- |
- |
hypothetical protein POPTR_0018s10190g [Populus trichocarpa] |