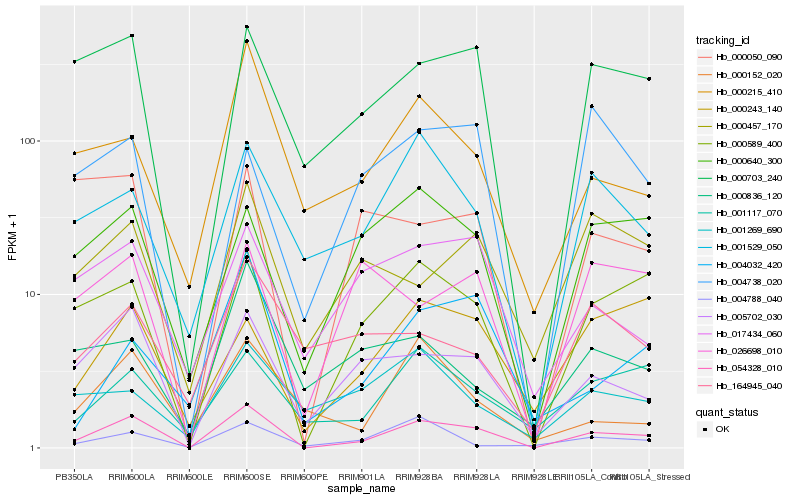
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054328_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0019s09820g [Populus trichocarpa] |
| 2 |
Hb_000589_400 |
0.1819419386 |
- |
- |
ESC, putative [Ricinus communis] |
| 3 |
Hb_005702_030 |
0.2035723369 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X2 [Vitis vinifera] |
| 4 |
Hb_000703_240 |
0.2125896778 |
- |
- |
- |
| 5 |
Hb_000640_300 |
0.2330472771 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 6 |
Hb_001529_050 |
0.2404715361 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 7 |
Hb_000215_410 |
0.2406929074 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 8 |
Hb_001117_070 |
0.2442044988 |
- |
- |
30S ribosomal protein S19, putative [Ricinus communis] |
| 9 |
Hb_017434_060 |
0.2484416265 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 10 |
Hb_000050_090 |
0.2550125664 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 11 |
Hb_000457_170 |
0.2559156024 |
- |
- |
PREDICTED: uncharacterized protein LOC105633066 [Jatropha curcas] |
| 12 |
Hb_000243_140 |
0.257159143 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 13 |
Hb_004032_420 |
0.2576567342 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 25-like [Jatropha curcas] |
| 14 |
Hb_000152_020 |
0.2586139606 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH118-like [Jatropha curcas] |
| 15 |
Hb_026698_010 |
0.2648911818 |
- |
- |
2,4-dienoyl-CoA reductase, putative [Ricinus communis] |
| 16 |
Hb_004788_040 |
0.2719256328 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_004738_020 |
0.2743404468 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000836_120 |
0.2756372896 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 19 |
Hb_164945_040 |
0.2781155139 |
- |
- |
PREDICTED: uncharacterized protein LOC105628261 [Jatropha curcas] |
| 20 |
Hb_001269_690 |
0.278581293 |
- |
- |
conserved hypothetical protein [Ricinus communis] |