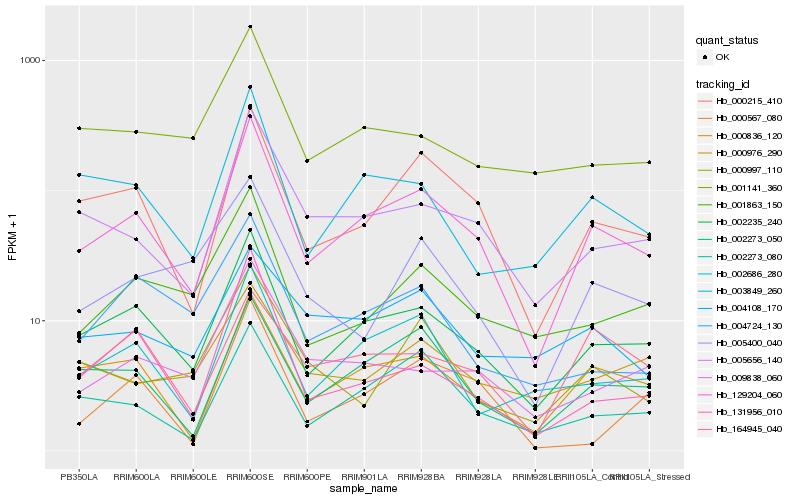
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_129204_060 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 2 |
Hb_002235_240 |
0.084129084 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 3 |
Hb_000215_410 |
0.1167512881 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 4 |
Hb_000836_120 |
0.1250704694 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 5 |
Hb_002273_050 |
0.1395211895 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_005656_140 |
0.1414263744 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 7 |
Hb_002686_280 |
0.1537647449 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_003849_260 |
0.1547208185 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 9 |
Hb_002273_080 |
0.1560637 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 10 |
Hb_001863_150 |
0.1669093701 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 11 |
Hb_009838_060 |
0.1726091246 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001141_360 |
0.1820304459 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 13 |
Hb_131956_010 |
0.1863546148 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 14 |
Hb_004724_130 |
0.187357743 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 15 |
Hb_004108_170 |
0.1900902135 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_005400_040 |
0.1904873011 |
- |
- |
PREDICTED: uncharacterized protein At4g04775-like [Jatropha curcas] |
| 17 |
Hb_000567_080 |
0.191672782 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 18 |
Hb_164945_040 |
0.1929296931 |
- |
- |
PREDICTED: uncharacterized protein LOC105628261 [Jatropha curcas] |
| 19 |
Hb_000976_290 |
0.1958250999 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 20 |
Hb_000997_110 |
0.1972853587 |
- |
- |
PREDICTED: uncharacterized protein LOC105632092 [Jatropha curcas] |