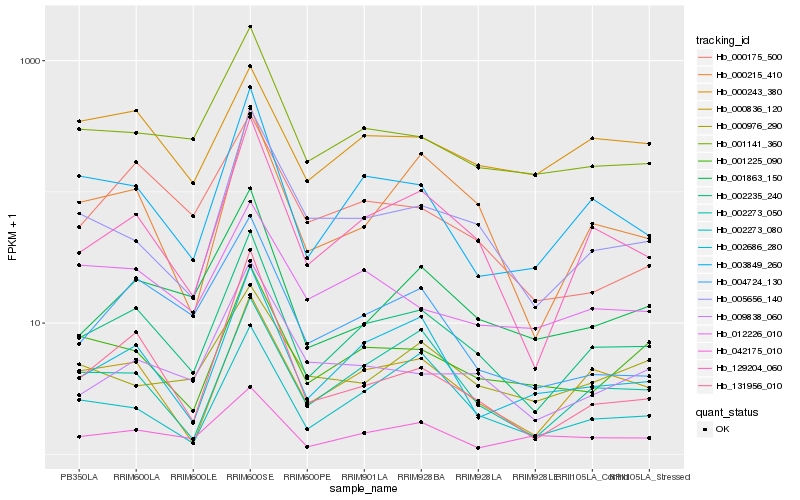
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_240 |
0.0 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 2 |
Hb_129204_060 |
0.084129084 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 3 |
Hb_000836_120 |
0.1169057477 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_003849_260 |
0.1195266308 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 5 |
Hb_000215_410 |
0.1353019552 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 6 |
Hb_001141_360 |
0.1384344211 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 7 |
Hb_001863_150 |
0.1390113241 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 8 |
Hb_002686_280 |
0.1408177959 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_002273_050 |
0.1471753073 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001225_090 |
0.1481354771 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_005656_140 |
0.1492785832 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 12 |
Hb_004724_130 |
0.1533549461 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14 [Jatropha curcas] |
| 13 |
Hb_009838_060 |
0.1594188116 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_042175_010 |
0.1602411001 |
- |
- |
hypothetical protein CISIN_1g0016721mg, partial [Citrus sinensis] |
| 15 |
Hb_000175_500 |
0.1656425655 |
- |
- |
PREDICTED: uncharacterized protein At1g15400 [Populus euphratica] |
| 16 |
Hb_002273_080 |
0.1686632186 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 17 |
Hb_131956_010 |
0.1700605481 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 18 |
Hb_012226_010 |
0.172078797 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 19 |
Hb_000976_290 |
0.1745786829 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 20 |
Hb_000243_380 |
0.1787582232 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |