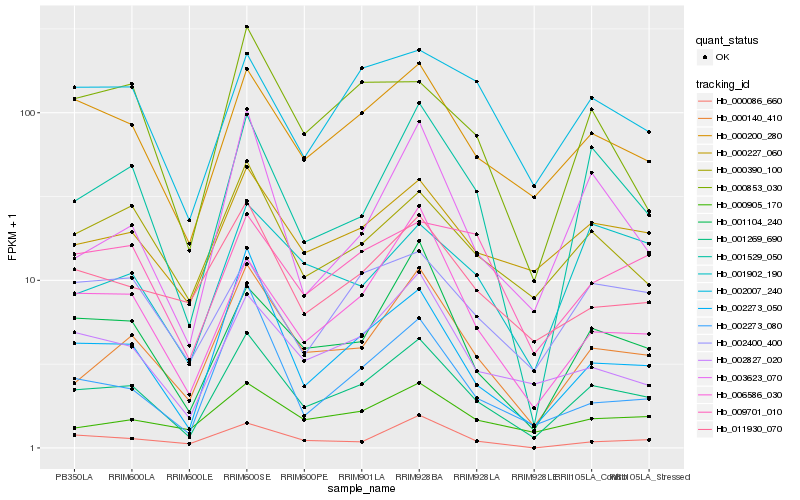
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_690 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000140_410 |
0.1081104244 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000200_280 |
0.1139264269 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 4 |
Hb_001529_050 |
0.123497946 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 5 |
Hb_002273_050 |
0.12358388 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000227_060 |
0.1326601752 |
transcription factor |
TF Family: NF-YC |
hypothetical protein POPTR_0010s03730g [Populus trichocarpa] |
| 7 |
Hb_001104_240 |
0.1332912886 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_002827_020 |
0.1364065016 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 9 |
Hb_000390_100 |
0.1372824877 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 10 |
Hb_000905_170 |
0.1433666942 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 11 |
Hb_011930_070 |
0.1448570328 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 12 |
Hb_006586_030 |
0.1460417332 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 13 |
Hb_002007_240 |
0.1531770041 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 14 |
Hb_002400_400 |
0.1549301078 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X3 [Jatropha curcas] |
| 15 |
Hb_003623_070 |
0.156539409 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 16 |
Hb_000853_030 |
0.157365756 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 17 |
Hb_000086_660 |
0.1591744417 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 18 |
Hb_002273_080 |
0.1611038877 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 19 |
Hb_009701_010 |
0.1639114731 |
- |
- |
PREDICTED: putative inactive cadmium/zinc-transporting ATPase HMA3 [Jatropha curcas] |
| 20 |
Hb_001902_190 |
0.1654200925 |
- |
- |
PREDICTED: oxalate--CoA ligase [Jatropha curcas] |