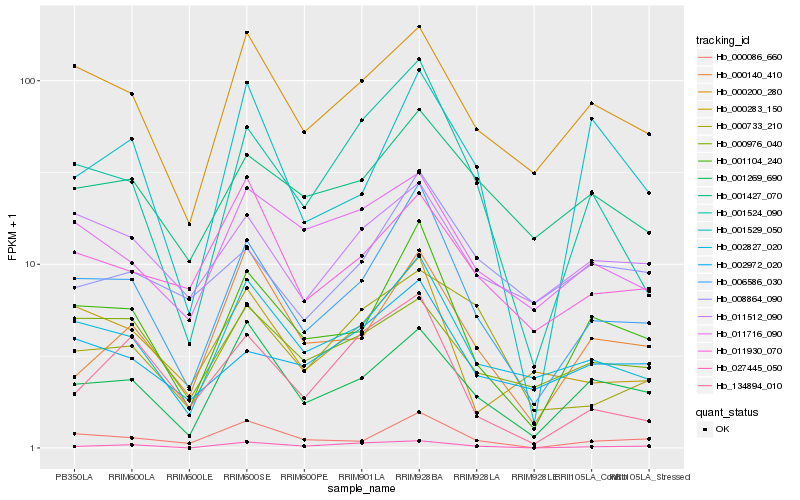
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006586_030 |
0.0 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 2 |
Hb_001104_240 |
0.0870798435 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_002827_020 |
0.1152271404 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 4 |
Hb_001524_090 |
0.1308260247 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |
| 5 |
Hb_000086_660 |
0.1393704677 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 6 |
Hb_001269_690 |
0.1460417332 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000200_280 |
0.151273387 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 8 |
Hb_000283_150 |
0.1649207486 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 9 |
Hb_002972_020 |
0.1688071892 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 10 |
Hb_001427_070 |
0.1737020267 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 11 |
Hb_000140_410 |
0.1749705236 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 12 |
Hb_011716_090 |
0.1797937041 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 13 |
Hb_011512_090 |
0.1797997882 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 14 |
Hb_000976_040 |
0.1838184005 |
- |
- |
molybdopterin cofactor sulfurase, putative [Ricinus communis] |
| 15 |
Hb_001529_050 |
0.1842119921 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 16 |
Hb_000733_210 |
0.1842290359 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 17 |
Hb_134894_010 |
0.1852644984 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 18 |
Hb_011930_070 |
0.1875170736 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 19 |
Hb_027445_050 |
0.1883435385 |
- |
- |
PREDICTED: titin isoform X13 [Jatropha curcas] |
| 20 |
Hb_008864_090 |
0.1902232295 |
- |
- |
PREDICTED: plastid division protein PDV2-like [Jatropha curcas] |