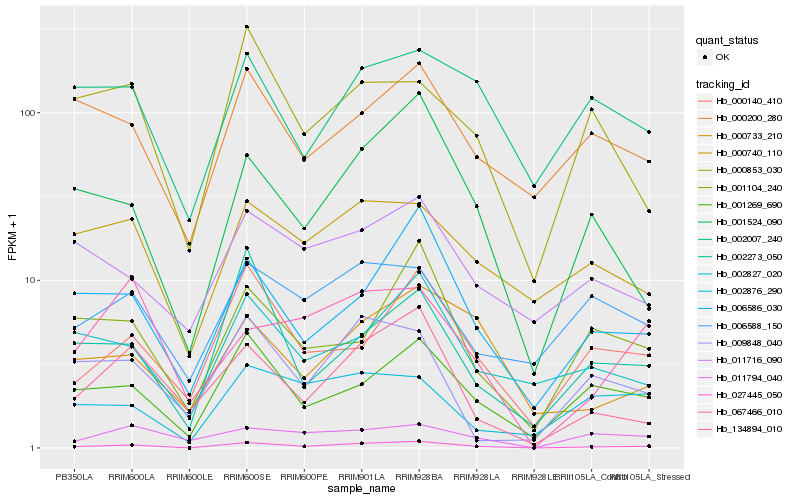
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027445_050 |
0.0 |
- |
- |
PREDICTED: titin isoform X13 [Jatropha curcas] |
| 2 |
Hb_001269_690 |
0.1788423243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001524_090 |
0.1873516873 |
- |
- |
PREDICTED: uncharacterized protein LOC105113929 [Populus euphratica] |
| 4 |
Hb_006586_030 |
0.1883435385 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 5 |
Hb_134894_010 |
0.1960656412 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 6 |
Hb_000140_410 |
0.1979610593 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000733_210 |
0.2115539943 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 8 |
Hb_002827_020 |
0.2127309705 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 9 |
Hb_002273_050 |
0.2135526881 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001104_240 |
0.2138046728 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 11 |
Hb_000740_110 |
0.2188995742 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_01850 [Jatropha curcas] |
| 12 |
Hb_000200_280 |
0.223786909 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 13 |
Hb_006588_150 |
0.2255221072 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 14 |
Hb_000853_030 |
0.2260912263 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 15 |
Hb_009848_040 |
0.2265840712 |
- |
- |
hypothetical protein B456_009G349600 [Gossypium raimondii] |
| 16 |
Hb_002007_240 |
0.2273582446 |
- |
- |
PREDICTED: F-box protein At1g55000 [Jatropha curcas] |
| 17 |
Hb_002876_290 |
0.227518781 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 18 |
Hb_011716_090 |
0.2312111054 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 19 |
Hb_067466_010 |
0.2313869942 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 20 |
Hb_011794_040 |
0.2315683519 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |