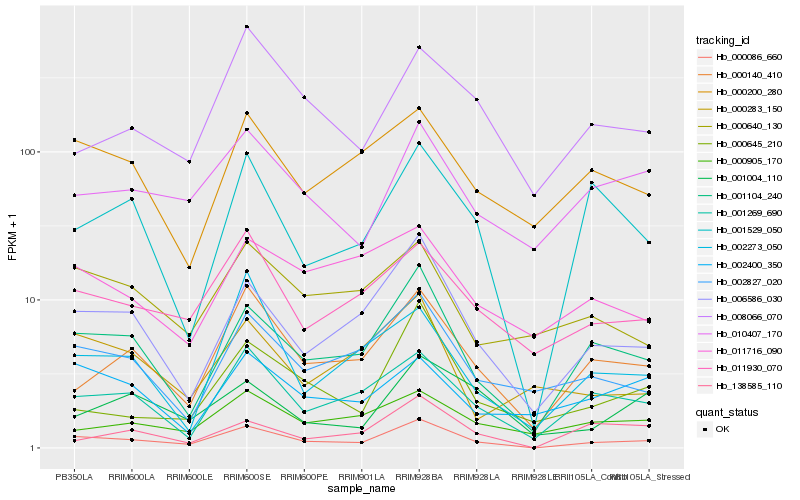
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_660 |
0.0 |
- |
- |
PREDICTED: phytosulfokine receptor 1-like [Jatropha curcas] |
| 2 |
Hb_001104_240 |
0.1254126509 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_006586_030 |
0.1393704677 |
- |
- |
PREDICTED: transcription factor LHW isoform X1 [Jatropha curcas] |
| 4 |
Hb_001269_690 |
0.1591744417 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000140_410 |
0.160230452 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 6 |
Hb_011930_070 |
0.1759474484 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 7 |
Hb_002827_020 |
0.1795935936 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 5-like [Jatropha curcas] |
| 8 |
Hb_001529_050 |
0.1799203837 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 9 |
Hb_010407_170 |
0.1853232868 |
- |
- |
PREDICTED: actin-depolymerizing factor-like [Jatropha curcas] |
| 10 |
Hb_000200_280 |
0.1937670329 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 11 |
Hb_000645_210 |
0.199531442 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_008066_070 |
0.2006760931 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 13 |
Hb_000640_130 |
0.2053062001 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_000905_170 |
0.2055996454 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 15 |
Hb_138585_110 |
0.2069411287 |
- |
- |
- |
| 16 |
Hb_002273_050 |
0.2074298957 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_001004_110 |
0.2128968265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011716_090 |
0.2140052539 |
transcription factor |
TF Family: HRT |
putative transciption factor family protein [Populus trichocarpa] |
| 19 |
Hb_002400_350 |
0.214022415 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 20 |
Hb_000283_150 |
0.2149655555 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |