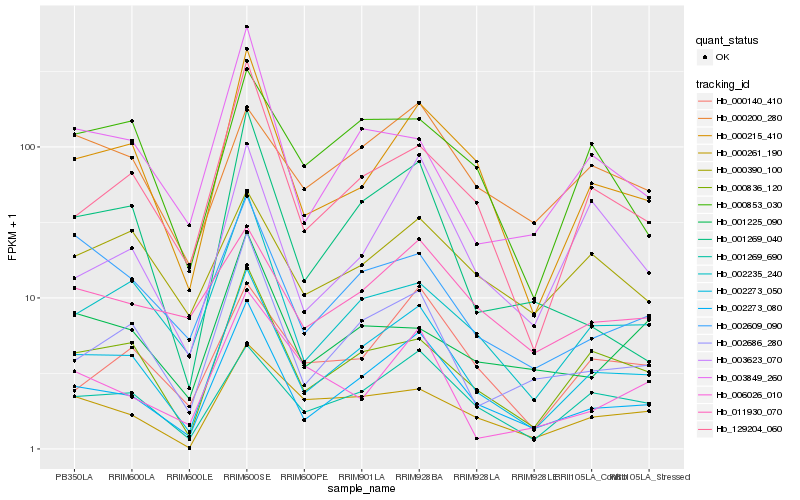
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002273_050 |
0.0 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_002273_080 |
0.0749684892 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 3 |
Hb_000215_410 |
0.1132030514 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 4 |
Hb_000836_120 |
0.1153202979 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 5 |
Hb_001269_690 |
0.12358388 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002686_280 |
0.1287801255 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_129204_060 |
0.1395211895 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 8 |
Hb_002235_240 |
0.1471753073 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 9 |
Hb_000261_190 |
0.1538143584 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Jatropha curcas] |
| 10 |
Hb_000140_410 |
0.1601986815 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002609_090 |
0.1666824738 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 12 |
Hb_003623_070 |
0.1698918592 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000853_030 |
0.1738180846 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 14 |
Hb_003849_260 |
0.1765340143 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 15 |
Hb_001269_040 |
0.1775018021 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_011930_070 |
0.178876491 |
- |
- |
PREDICTED: probable AMP deaminase [Jatropha curcas] |
| 17 |
Hb_000390_100 |
0.1857166818 |
- |
- |
alpha-hydroxynitrile lyase family protein [Populus trichocarpa] |
| 18 |
Hb_006026_010 |
0.1871521807 |
- |
- |
PREDICTED: cyclin-dependent kinase inhibitor 3-like [Jatropha curcas] |
| 19 |
Hb_000200_280 |
0.1882329881 |
- |
- |
PREDICTED: uncharacterized protein LOC105636924 [Jatropha curcas] |
| 20 |
Hb_001225_090 |
0.1883587248 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |