| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
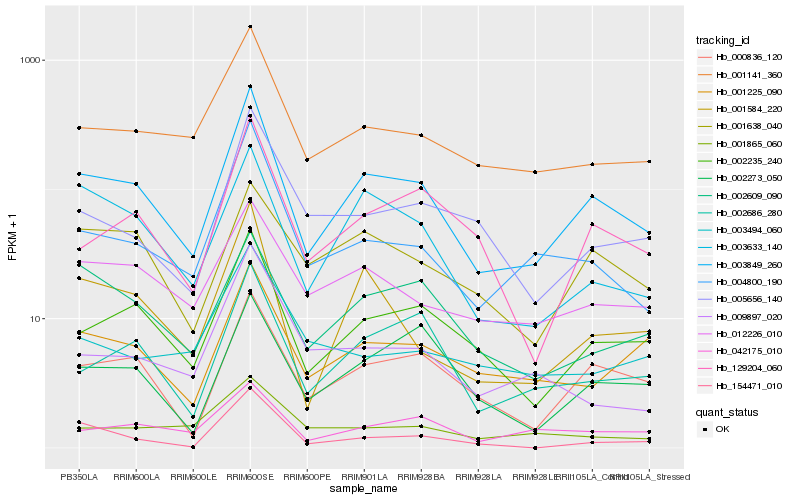
Hb_003849_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644666 [Jatropha curcas] |
| 2 |
Hb_002235_240 |
0.1195266308 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 3 |
Hb_000836_120 |
0.1324261761 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_001141_360 |
0.1426245063 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 5 |
Hb_004800_190 |
0.1434621466 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 4 [Jatropha curcas] |
| 6 |
Hb_001584_220 |
0.1482991265 |
- |
- |
galactinol synthase [Manihot esculenta] |
| 7 |
Hb_002686_280 |
0.1527771374 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_129204_060 |
0.1547208185 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 9 |
Hb_001225_090 |
0.1591597885 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_012226_010 |
0.1607560926 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 11 |
Hb_003633_140 |
0.1609035288 |
- |
- |
PREDICTED: uncharacterized protein LOC105647486 isoform X1 [Jatropha curcas] |
| 12 |
Hb_042175_010 |
0.1656403652 |
- |
- |
hypothetical protein CISIN_1g0016721mg, partial [Citrus sinensis] |
| 13 |
Hb_002609_090 |
0.171715885 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_005656_140 |
0.1717728485 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 15 |
Hb_002273_050 |
0.1765340143 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_001638_040 |
0.1794861389 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 17 |
Hb_154471_010 |
0.1867573647 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 18 |
Hb_009897_020 |
0.1892706594 |
- |
- |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 19 |
Hb_001865_060 |
0.1904104988 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 20 |
Hb_003494_060 |
0.1930455188 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |