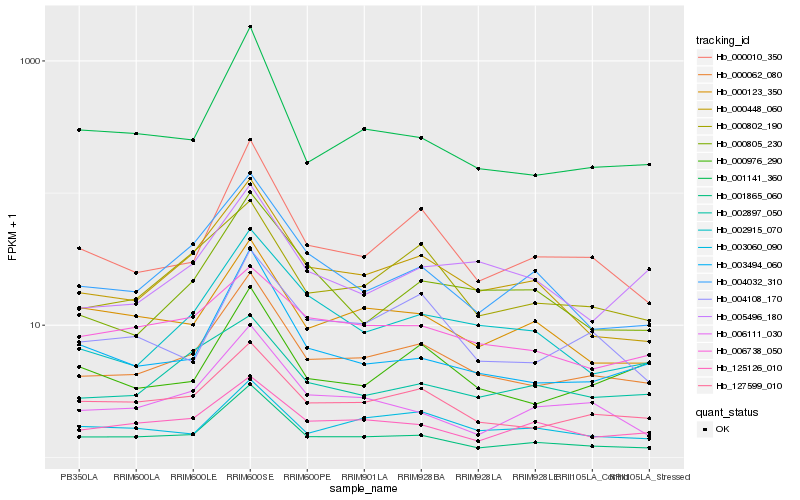
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_080 |
0.0 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR11-like [Jatropha curcas] |
| 2 |
Hb_001865_060 |
0.0924008613 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 3 |
Hb_127599_010 |
0.0935141085 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 4 |
Hb_000448_060 |
0.1035269842 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 5 |
Hb_000802_190 |
0.1053304058 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 6 |
Hb_000010_350 |
0.1067472084 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 7 |
Hb_001141_360 |
0.113622377 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 8 |
Hb_000976_290 |
0.1149273979 |
transcription factor |
TF Family: MYB-related |
PREDICTED: uncharacterized protein LOC105646228 [Jatropha curcas] |
| 9 |
Hb_002915_070 |
0.1197929456 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 10 |
Hb_003494_060 |
0.119997884 |
- |
- |
PREDICTED: uncharacterized protein LOC105644062 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003060_090 |
0.1255860733 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 12 |
Hb_004108_170 |
0.1266219827 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005496_180 |
0.1277448146 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 14 |
Hb_004032_310 |
0.1301492031 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 15 |
Hb_125126_010 |
0.1303438777 |
desease resistance |
Gene Name: NB-ARC |
resistance family protein [Populus trichocarpa] |
| 16 |
Hb_000123_350 |
0.1304036566 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_000805_230 |
0.1307748953 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 18 |
Hb_006111_030 |
0.1341211871 |
- |
- |
PREDICTED: phospholipase A(1) LCAT3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002897_050 |
0.1342481892 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 20 |
Hb_006738_050 |
0.1343276722 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |