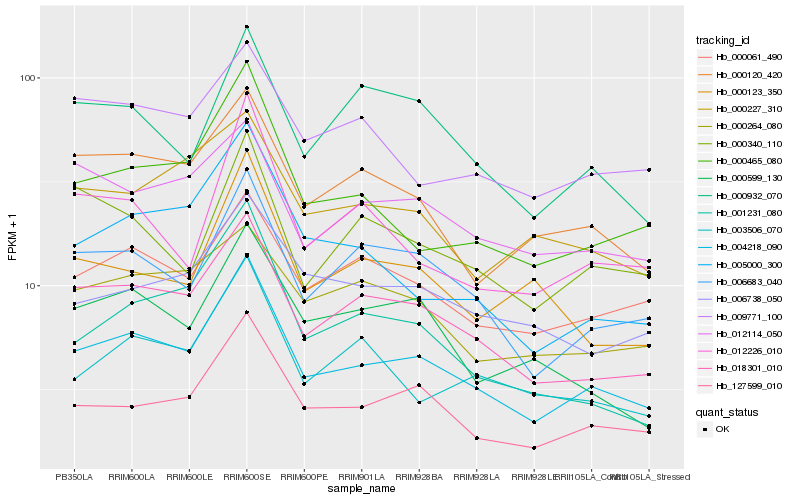
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_090 |
0.0 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_018301_010 |
0.0749475666 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Jatropha curcas] |
| 3 |
Hb_127599_010 |
0.0772548417 |
- |
- |
hypothetical protein CISIN_1g0010462mg, partial [Citrus sinensis] |
| 4 |
Hb_006683_040 |
0.0910871642 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 5 |
Hb_000465_080 |
0.0986318904 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 6 |
Hb_000061_490 |
0.1018235104 |
- |
- |
hypothetical protein CISIN_1g0362002mg, partial [Citrus sinensis] |
| 7 |
Hb_000120_420 |
0.1043377682 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5-like [Jatropha curcas] |
| 8 |
Hb_005000_300 |
0.1131361468 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 9 |
Hb_003506_070 |
0.1145174496 |
- |
- |
PREDICTED: calcineurin B-like protein 7 isoform X2 [Jatropha curcas] |
| 10 |
Hb_006738_050 |
0.1149996106 |
- |
- |
PREDICTED: uncharacterized protein LOC105634992 [Jatropha curcas] |
| 11 |
Hb_012114_050 |
0.1151813522 |
- |
- |
PREDICTED: uncharacterized protein LOC105645815 isoform X1 [Jatropha curcas] |
| 12 |
Hb_012226_010 |
0.1168395321 |
- |
- |
PREDICTED: protein DGCR14 [Jatropha curcas] |
| 13 |
Hb_000932_070 |
0.1172724144 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 14 |
Hb_000340_110 |
0.1174557256 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 15 |
Hb_009771_100 |
0.119094882 |
- |
- |
PREDICTED: uncharacterized protein LOC105141784 [Populus euphratica] |
| 16 |
Hb_000599_130 |
0.1192162002 |
- |
- |
PREDICTED: U-box domain-containing protein 3 [Jatropha curcas] |
| 17 |
Hb_000264_080 |
0.1205174836 |
- |
- |
hypothetical protein JCGZ_21412 [Jatropha curcas] |
| 18 |
Hb_000123_350 |
0.1206771775 |
desease resistance |
Gene Name: ABC_membrane |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_001231_080 |
0.1207813085 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like isoform X1 [Fragaria vesca subsp. vesca] |
| 20 |
Hb_000227_310 |
0.1208455978 |
- |
- |
PREDICTED: cyclin-T1-4-like isoform X2 [Jatropha curcas] |