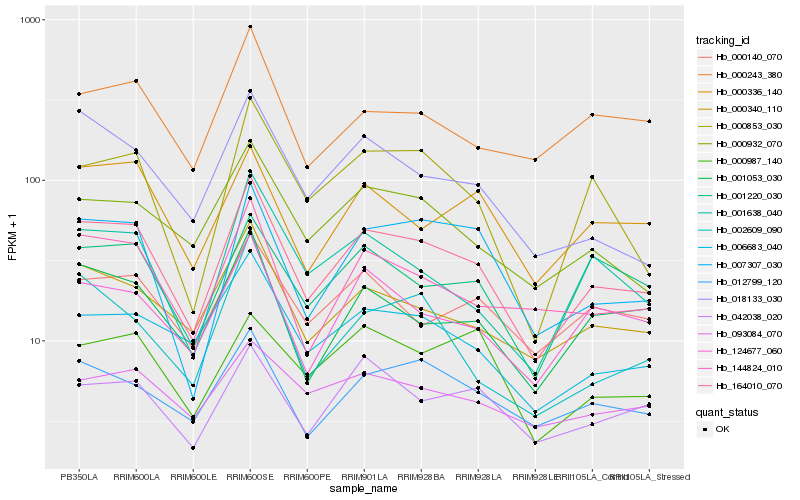
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164010_070 |
0.0 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 2 |
Hb_000340_110 |
0.0987651041 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 3 |
Hb_144824_010 |
0.104346011 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |
| 4 |
Hb_124677_060 |
0.1050962081 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_006683_040 |
0.1058148006 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 6 |
Hb_001053_030 |
0.1068051087 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000932_070 |
0.1075871768 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 8 |
Hb_007307_030 |
0.1088849118 |
- |
- |
PREDICTED: myosin-11 isoform X2 [Jatropha curcas] |
| 9 |
Hb_012799_120 |
0.1126877986 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001638_040 |
0.1136037839 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 11 |
Hb_018133_030 |
0.118494803 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 12 |
Hb_000336_140 |
0.1185275232 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000140_070 |
0.1189893014 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 14 |
Hb_042038_020 |
0.12003496 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 15 |
Hb_000853_030 |
0.1214732386 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 16 |
Hb_001220_030 |
0.1219151496 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 17 |
Hb_000243_380 |
0.1235605689 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 18 |
Hb_093084_070 |
0.132865935 |
desease resistance |
Gene Name: Baculo_PEP_C |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 19 |
Hb_002609_090 |
0.1329231233 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 20 |
Hb_000987_140 |
0.1349036301 |
- |
- |
PREDICTED: uncharacterized protein LOC105648083 isoform X1 [Jatropha curcas] |