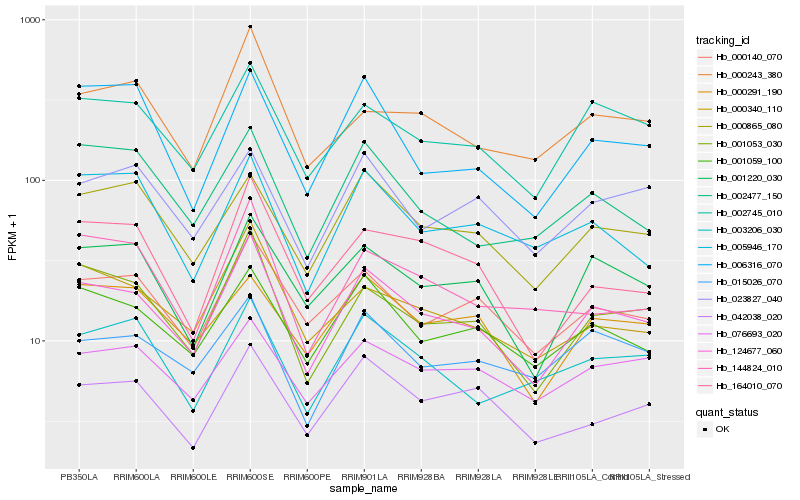
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124677_060 |
0.0 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_001053_030 |
0.0755653562 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_002745_010 |
0.0878746168 |
- |
- |
PREDICTED: SUMO-conjugating enzyme SCE1 [Cucumis sativus] |
| 4 |
Hb_001059_100 |
0.0933221385 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein U-like protein 1 [Jatropha curcas] |
| 5 |
Hb_000140_070 |
0.0959781873 |
- |
- |
PREDICTED: uncharacterized protein LOC105642935 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001220_030 |
0.0970155124 |
- |
- |
PREDICTED: BRCA1-A complex subunit Abraxas [Jatropha curcas] |
| 7 |
Hb_000340_110 |
0.0972479631 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X1 [Jatropha curcas] |
| 8 |
Hb_000865_080 |
0.1042778834 |
- |
- |
arsenite-resistance protein, putative [Ricinus communis] |
| 9 |
Hb_164010_070 |
0.1050962081 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 10 |
Hb_042038_020 |
0.1070916219 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 11 |
Hb_015026_070 |
0.1083132688 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000243_380 |
0.1105787531 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 13 |
Hb_003206_030 |
0.1110963587 |
- |
- |
PREDICTED: uncharacterized protein LOC105632652 [Jatropha curcas] |
| 14 |
Hb_076693_020 |
0.1117612587 |
- |
- |
hypothetical protein CISIN_1g0121812mg, partial [Citrus sinensis] |
| 15 |
Hb_002477_150 |
0.1122047583 |
transcription factor |
TF Family: Alfin-like |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_144824_010 |
0.1137494538 |
- |
- |
hypothetical protein POPTR_0019s02110g [Populus trichocarpa] |
| 17 |
Hb_000291_190 |
0.1153865269 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA3 [Jatropha curcas] |
| 18 |
Hb_006316_070 |
0.1181621577 |
transcription factor |
TF Family: GeBP |
PREDICTED: mediator-associated protein 1-like [Jatropha curcas] |
| 19 |
Hb_005946_170 |
0.1185900769 |
- |
- |
PREDICTED: FRIGIDA-like protein 4a [Jatropha curcas] |
| 20 |
Hb_023827_040 |
0.1201389078 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |